Reconstructing the evolutionary history of cancer using artificial intelligence
Researchers from Barts Cancer Institute (BCI), Queen Mary University of London, and The Institute of Cancer Research, London have developed a computational model that can reconstruct the evolutionary history of cancer. By unravelling the genetic complexity of a tumour, the tool can be used to better understand how the cancer has developed and may help to guide treatment strategies in the future.
Combining machine learning and evolutionary theories
In the study, published today in Nature Genetics, the team created a computer model that combines artificial intelligence with the mathematical theories of Darwin’s evolution. By interpreting the genetic sequences of tumour samples, the model is able to find patterns of genetic faults that exist in the tumour and determine how these arose and accumulated over the course of the cancer’s development.
Professor Trevor Graham, lead researcher of the study from the BCI, said:
“Tumours evolve, and understanding their evolution is key to improving patient outcomes. Our work shows how interpreting the cancer genome through the lens of evolution, and with the help of artificial intelligence, gives us a much more accurate picture of how tumours change over time than was previously available.”
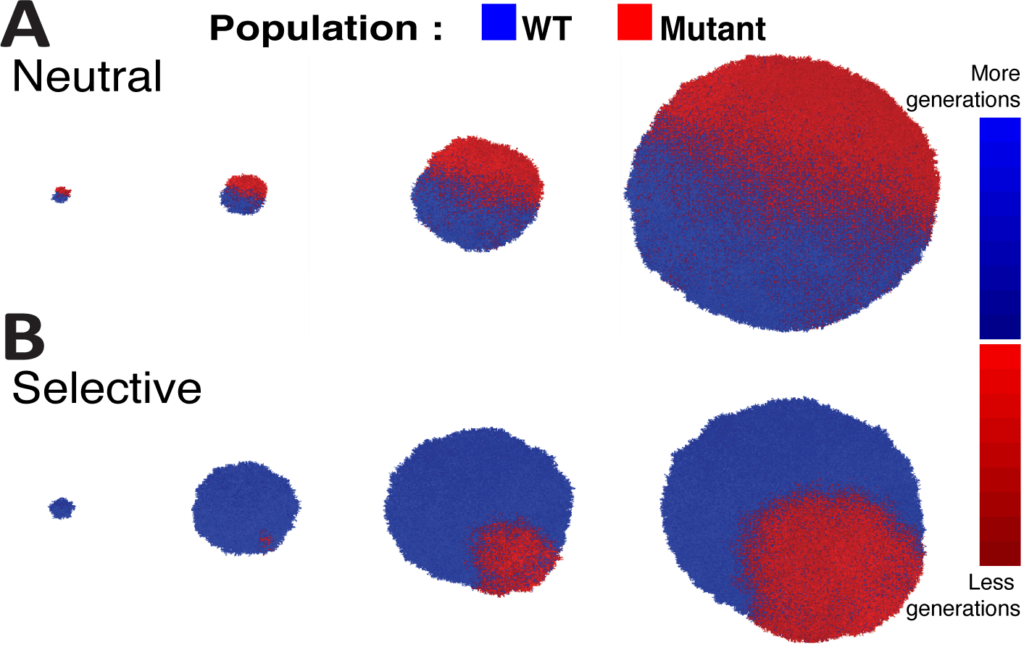
During cancer development, different cells acquire different faults within their DNA, known as mutations. As the tumour continues to grow and become more advanced, the cells replicate and create more cells with the same mutation. This process generates genetically diverse tumours made up of distinct populations of cells called ‘subclones.’
By teaching the computer model the theories of evolution and applying these new algorithms to genetic data, the team found that tumours actually had a simpler genetic structure than previously thought; with a third to a quarter fewer subclones than research has previously suggested. The model could also determine how old each subclone was and how fast it was growing.
Professor Andrea Sottoriva, lead researcher of the study from The Institute of Cancer Research (ICR), said:
“Tumours are a microcosm of evolution, with different populations of cells competing to survive and responding to environmental pressures. By harnessing the computational power of artificial intelligence and combining it with evolutionary theories developed since Darwin’s time, our models were able to make sense of the complex genetic data and accurately reconstruct the tumour’s genetic history.”
The team applied their model to genetic data from thousands of cancer samples, including samples of breast cancer, bowel cancer, leukaemia and brain tumours.
The research was primarily funded by the Wellcome Trust and Cancer Research UK, with additional funding from the Medical Research Council, the National Institute of Health and Children with Cancer UK.
Eliminating the background noise
Although numerous mutations accumulate within tumour cells, not all of them are important for a cancer’s development. These neutral or ‘hitchhiker’ mutations, which provide neither an advantage nor disadvantage to the cancer, build up randomly within certain cells of a tumour in a process called ‘neutral evolution.’ Previous computational tools have failed to take neutral evolution into account, making it more difficult to identify important mutations that have contributed to a cancer’s development.
By cutting out the background noise generated by neutral evolution, this new model generates a more accurate reading of the genetic data, making it easier to identify key mutations, such as those that drive cancer progression, spread or treatment resistance.
Informing treatment decisions
The Genomic Medicine initiative within the NHS means that the use of cancer genome data is set to become a routine part of patient care, and having the tools in place to accurately interpret these data to benefit patients is very important.
In the future, the tool may be able to be applied in clinical practise to guide treatment strategies. By identifying what mutations are key for driving a particular cancer, more informed decisions can be made on what treatment may work best. Knowing the pattern of mutations that exist in a tumour may also offer some indication of what mutations are likely to come next, providing the opportunity to administer the most effective treatment in light of the prediction.
The tool offers a powerful window into the cancer genome, and the team hope that, in time, their tool may help to better understand and treat cancers to improve outcomes for patients.
Category: General News, Publications

No comments yet