MRC Doctoral Training Partnership Programme
in Translational Biomedical Sciences
Overview
Queen Mary University of London and the University of Southampton are proud to run a joint Medical Research Council (MRC)-funded Doctoral Training Partnership. Starting in 2016 in collaboration with the MRC as "MRC DTP in Translational Immunology, Inflammation and Cancer", we have established a successful 4-year MRC-funded MRes/PhD programme that identifies excellent students and matches them with outstanding research projects. After the successful renewal, the programme was renamed to "MRC DTP in Translational Biomedical Sciences” in September 2022, incorporating the themes of cancer, neurological, inflammatory and infectious diseases, anti-microbial resistance and global health.
Recently we also appointed additional MRC-funded students to study projects addressing skills of national shortage and iCASE studentships who study in association with a commercial partner. Together with university funding, the joint programmes have appointed over 60 students to our MRC DTP to date. Our students experience a variety of research environments and skills that arm them for their future career in the field of science. We are enormously proud of our students, many of whom offer commentary on their experiences to date.
All studentships are fully funded including 4 years of stipend (equivalent to the Research Councils UK National Minimum Doctoral Stipend) and fees, as well as some research funds.
There are two types of studentships available - MRes/PhD and iCASE:
MRes/PhD
The programme is 1 year MRes + 3 years PhD. The MRes/PhD is undertaken at either the Faculty of Medicine and Dentistry, Queen Mary or the Faculty of Medicine, University of Southampton. In the MRes year (year 1) students study (at either Queen Mary or Southampton) three taught modules that include statistics, computational coding and quantitative biology skills. Additionally, students undertake three 12-week rotation projects (usually two wet-lab studies and one of which must be predominantly mathematics, computational biology or bioinformatics) which are each assessed by a mini-thesis report. On successful completion and award of the MRes, students select one of the projects as the subject of their 3-year PhD (year 2-4).
iCASE
iCASE (industrial Collaborative Awards in Science and Engineering) PhD projects are developed and carried out during the four-year PhD in close collaboration with a non-academic partner. The iCASE PhD is undertaken at either the Faculty of Medicine and Dentistry, Queen Mary or the Faculty of Medicine, University of Southampton. Within your PhD you will spend a period of time with the non-academic partner (usually three to six months over the lifetime of the PhD).
Students, who choose the iCASE studentship may select their PhD project from year one, i.e., they don’t undertake the three 12-week rotation projects. Furthermore, participation in the three taught modules is voluntary and not compulsory as in the MRes.


Funded by

Charterhouse Square Campus, Queen Mary University of London

Faculty of Medicine, University of Southampton

Meet the students
Cohort 2022
Disha Mehta (Southampton)
Cancer Immunology
Which rotation did you decide to pursue for your PhD project and why did you choose it? My PhD research is focussed on understanding T cell exhaustion within the context of immunotherapy. Prior to selecting my rotations last year, I had a predetermined project offer that allowed me to explore rotations in my supervisor's labs. This approach aimed to provide me with a better understanding of their lab operations and foster closer relationships with them. I chose three rotations, each offering distinct skill sets ranging from bioinformatics coding and data analysis to in vitro wet lab techniques. The skills I acquired during these rotations significantly contributed to my final PhD project, particularly the first two rotations that involved extensive single-cell RNA sequencing analysis and bioinformatic trajectory inference analysis. These experiences have given me a head start in my first year of data collection.
What skills have you developed so far? All three rotations provided me with the opportunity to acquire new skills in Bioinformatics, including programming in R and conducting single-cell RNA sequencing analysis on datasets sourced from human patient samples. Additionally, I developed a novel tool designed to facilitate the understanding of gene-switching patterns along defined trajectories. In the wet lab, I gained proficiency in techniques such as Western blotting and flow cytometry, as well as in vitro methods like T cell proliferation assays and shRNA knockdown experiments.
During my first year, my organizational skills significantly improved as I learned to maintain a detailed and up-to-date lab notebook, and I became adept at preparing thoroughly before conducting experiments. Furthermore, my communication and interpersonal abilities underwent substantial development through numerous interactions with individuals in different labs. I also honed my presentation skills, confidently sharing my progress in weekly lab meetings, away day events, and various seminars.
How have you found the mathematic/computational component of the DTP? I have found the statistics module extremely useful and thoroughly detailed which has made it extremely easy to comprehend and apply different descriptive statistical tests using SPSS and GraphPad PRISM to plot my data efficiently. The computational component of the DTP has been the most useful as my project heavily relies on single cell RNA sequencing and I have enjoyed the practical sessions which has given me the opportunity to translate the theoretical knowledge onto actual datasets.
What have you enjoyed most about your experience so far? After completing an intense year, I've come to the realization that I thoroughly enjoyed every aspect of engaging in three different projects and acquiring new skills both in the lab and in dry lab analysis. The experience of learning something new every day, improving my presentation skills, and gaining the confidence to work independently was incredibly rewarding. Prior to this, I had no experience with R programming, but the structured quantitative computational component of the DTP facilitated my learning process, especially in areas like single-cell RNA sequencing, with interactive teamwork sessions enhancing my understanding.
The DTP has provided excellent opportunities, including online and in-person seminars and academic talks, which I found immensely helpful for my background research. I also appreciated the social events organized by the LSPS society, offering a fantastic platform to connect with fellow PhD students. Lastly, I am deeply grateful for the student-led away, particularly the media training sessions, which were truly insightful and the motivational talks left a lasting impression on me.
Ester Valero Hernández (Queen Mary)
Cancer, Inflammation and Immunology

Which rotation did you decide to pursue for your PhD project and why did you choose it? For my PhD, I decided to pursue my second rotation, which I took in Prof Angray Kang’s lab in the Blizard Institute, in close collaboration with Prof David Baker. The aim of my PhD project is to investigate the mechanisms of autoimmunity induced by Epstein Barr virus, and how it might be related to multiple sclerosis. I chose this project because I found the topic to be highly exciting and of high importance, keeping up to the date to the state of current research, and the translational potential of the project made it very attractive to me.
What skills have you developed so far? During the MRes year I undertook three very different rotations in order to make the most of my experience. As such, I further developed my lab skills using techniques such as cell culture, PCR and qPCR, immunohistochemistry and immunofluorescence of cell cultures and animal tissue, cryosectioning, bacteria transformation and protein purification, and more.
How have you found the mathematic/computational component of the DTP? For me, the computational component has definitely been the most challenging aspect of the programme so far. The R data carpentry sessions they introduced this year were incredibly helpful to set the basics, but the module can be quite time consuming if you haven’t got a strong computational background. However, the practical workshops are well organised, and the module organisers are incredibly helpful and willing to solve any issues and answer questions.
What have you enjoyed most about your experience so far? What have you found challenging My favourite part about the programme so far has been the rotations taken during the MRes year. I especially value the opportunity to reach out to investigators of our choice and discuss the possibility of designing a new study based on your particular interests; that is how I found my perfect project!
So far, the most challenging aspect has probably been balancing the workload of the rotations and taught modules altogether, but it definitely gets easier as you advance through the year.
Andrea Vinaga (Queen Mary)
Cancer, Inflammation and Immunology

What is your current rotation? My current rotation is in Dr Jun Wang's group at the Barts Cancer Institute, where I am using bulk and single cell RNA transcriptomics to study oesophageal squamous cell carcinoma. More specifically, I am trying to identify master transcription factors that may influence the tumour immune microenvironment.
What skills have you developed so far, and what skills do you hope to develop? My current project is fully bioinformatics, which I had not had much experience in before starting this programme. I have developed a range of skills in transcriptomic analyses, which have made me more confident in this area.
How have you found the mathematic/computational component of the DTP? I enjoyed the Quantitative Cell Biology module in Southampton, although travelling there every week can be tiring. The course covers every aspect of transcriptomics, from bulk to single cell RNAseq. The lecturers are very knowledgeable and happy to help with any issues you may encounter during the course. This module starts with a week-long R course, which I found extremely useful.
What have you enjoyed most about your experience so far? What have you found challenging? My favourite aspect of the programme is the opportunity to do three rotations, which allows you to find the best project and group fit for you. This also allows you to create useful networks for the course of your PhD. There are a lot of projects to choose from, but you can also reach out to other labs. The first rotation is the most coursework heavy, and I have found the workload stressful and hard to deal with at times.
Alex Robinson (Southampton)
Immunity and Infection
Which rotation did you decide to pursue for your PhD project and why did you choose it? I decided to merge my first and second rotations into a collaborative project between Professor Paul Elkington and Dr Liku Tezera of the Tuberculosis (TB) group, and Professor Tilman Sanchez-Elsner of the Junk DNA group. My project focuses on the role of extracellular vesicles (EVs) released by Mycobacterium tuberculosis infected cells in regulating inflammation in TB, with the ultimate aim of characterising these EVs for blood-based diagnostics, and even therapeutics, in TB.
What skills have you developed so far? Between the two labs, I have developed many skills, especially wet lab skills. I now have experience with techniques such as flow cytometry, qPCR, ELISAs, working in a category 3 lab. Alongside these are other basic wet lab skills such as planning experiments (from simple calculations and setups to more complex assays), time management, organisation, and develop my confidence in working independently. Through my third rotation, and with help from the Quantitative Cell Biology module, I have also greatly improved my skills in bioinformatics. I worked with large RNA-seq and scRNA-seq datasets in R and python, and used a wide variety of online bioinformatics tools.
How have you found the mathematic/computational component of the DTP? I have personally found the computational part of the DTP very interesting an enjoyable, opening a new area of bioinformatics to me which I previously thought was unattainable given I do not have a background in computing. This has translated into my third rotation project being a bioinformatic project, and even adding a bioinformatic element to my PhD research. I have also found the maths side of research to be one of the areas I struggle with, but there are plenty of opportunities to improve these skills through the introductory module and other courses on offer.
What have you enjoyed most about your experience so far? What have you found challenging? I have most enjoyed working in a collaborative lab environment here on Level E, meeting colleagues with very interesting research interests and making great friends in the process. The most challenging part of the PhD so far has been adjusting to the workload and expectation that a PhD demands, but it has been beneficial, and I can see the improvement in my quality of work as a result.
Liam Edgeway (Southampton)
Immunity and Infection
What rotation did you decide to pursue for your PhD project and why? I have joined Dr. Matthew Loxham’s group, where I will be undertaking a multidisciplinary approach to investigate the effects of airborne particulate matter on the human respiratory system, and how these effects may contribute to a disease state and an estimated 7 million deaths per year (WHO, 2018). I thoroughly enjoy using a range of methods and techniques when answering a question. The multidisciplinary approach in studying a major global health concern will not only allow for a varied and thorough understanding as to the composition and effects of particulate matter on the respiratory system, but will also help me to translate results to biological understanding, to guide policy, and potentially contribute to better health outcomes for the whole population.
What skills have you developed so far? The three rotations in the MRes year provided me with training in cell culture, fluorometric microassays, RT-qPCR, western blotting, ELISA, histological techniques, mass spectrometry, and bioinformatics. Additionally, the MRC DTP away day, regular lab meetings, and poster presentation sessions have provided me with a platform to enhance my presentation and communication skills.
How have you found the mathematic/computational component of the DTP? Having zero experience with bioinformatics before the DTP, the QCB module provided me with an in-depth theoretical understanding of many methods. This not only aided me in my decision-making when using bioinformatic techniques but also allowed me to understand data presented in the results sections of scientific papers.
The practical aspect of the QCB module also provided my first coding experience. A thorough understanding of the tools allowed me to build a solid foundation in R, and further provided the resources needed to improve these skills.
What have you enjoyed most about your experience so far? What have you found challenging? I joined the iPhD with a defined project, where I knew I would be staying in Dr. Loxham’s group following the MRes year. However, the rotation projects provided an opportunity to learn the subject area from different angles, with fresh approaches to tackling questions. I thoroughly enjoyed the variety of techniques and topics that were recruited over the year. The coursework can be a bit difficult to juggle at times, although, it certainly requires and develops time management and organisation!
Cohort 2021
Amy Hall (Queen Mary)
Cancer, Inflammation and Immunology
What is your current rotation? I am currently undertaking my first rotation with Dr Mirjana Efremova. This project focuses on the analysis of single cell RNA-sequencing (scRNA-Seq) data from patients with triple negative breast cancer, aiming to characterise the heterogenic cancer cell states and the interactions with the tumour microenvironment. This data is from treatment-naïve patients and can later be compared to mid- and post-treatment samples.
What skills have you developed so far? So far, I have been focusing on learning how to use R to analyse the scRNA-Seq data. I have no previous experience using R or analysing large data sets. Therefore, it has been interesting to learn how to use R and how it can be used to analyse patient data.
How have you found the mathematic/computational component of the DTP? The bioinformatics sessions in Southampton have been useful as learning R in these sessions has helped me with my current rotation. The sessions themselves can be difficult, especially when learning something new. However, the session leads are helpful and explain the concepts well.
What have you enjoyed most about your experience so far? What have you found challenging? The best part of this scheme is being able to join multiple labs during the MRes year, as it allows me to work with multiple supervisors but also to try things which I wouldn’t have the opportunity to do if I went straight into a PhD. I have least enjoyed balancing travelling weekly to Southampton, the research methods module and my first rotation all at once. The prep work and coursework for research methods is quite time-consuming when balancing it with everything else.
Natasha Palmer (Southampton)
Immunity and Infection
 What rotation did you decide to pursue for your PhD project and why? For my PhD project, I combined my last two rotations together, meaning I am part of a collaboration between laboratories run by Professor Tilman Sanchez-Elsner and Professor Salim Khakoo. My project focuses on the interaction between natural killer cells and macrophages in the tumour microenvironment and I ultimately aim to manipulate the relationship to boost anti-tumour responses. I ended up combining the rotations because I thoroughly enjoyed both and couldn’t decide on just one of them!
What rotation did you decide to pursue for your PhD project and why? For my PhD project, I combined my last two rotations together, meaning I am part of a collaboration between laboratories run by Professor Tilman Sanchez-Elsner and Professor Salim Khakoo. My project focuses on the interaction between natural killer cells and macrophages in the tumour microenvironment and I ultimately aim to manipulate the relationship to boost anti-tumour responses. I ended up combining the rotations because I thoroughly enjoyed both and couldn’t decide on just one of them!
What skills have you developed so far, and what skills do you hope to develop? I have developed useful laboratory skills in immunological research, such as immunohistochemistry techniques, ELISAs, qPCR, flow cytometry and tissue culture. I am now focusing on developing my time management skills and working independently in the laboratory, as well as improving my presentation skills.
How have you found the mathematic/computational component of the DTP? The bioinformatics module has been incredibly helpful as I have no previous experience in this area. It’s interesting to not only learn the techniques but to be given examples of how they are used in modern medical research. We have practical workshops to try out the coding and analysis and the demonstrators in these are very friendly and good at explaining everything.
What have you enjoyed most about your experience so far? What have you found challenging? Working between two research groups has been exciting and I have loved learning different techniques from each group. I have particularly enjoyed getting to grips with flow cytometry as it was quite daunting to begin with, but now I feel more confident and conquering a new technique is very satisfying. My main challenge has been coordinating space, resources and time between the two groups, for example it can be hard to manage your time during weeks when you are meeting both supervisors or attending two laboratory meetings.
Akash Saha (Queen Mary)
Cancer, Inflammation and Immunology
 What rotation did you choose for your PhD project and why? I decided to pursue my last rotation of the MRes year in Prof. Tyson Sharp’s lab. Here, my role is involved in characterising and targeting novel synthetically lethal vulnerabilities in LIMD1 deficient cancers.
What rotation did you choose for your PhD project and why? I decided to pursue my last rotation of the MRes year in Prof. Tyson Sharp’s lab. Here, my role is involved in characterising and targeting novel synthetically lethal vulnerabilities in LIMD1 deficient cancers.
I chose this project for several reasons, including the novelty of the work related to it since the main aim is to characterise exciting novel targets identified from two whole genome CRISPR functional screens performed in collaboration with the CRUK Functional Genomic Centre. As LIMD1 is a tumour suppressor gene frequently lost during the early stages of lung carcinogenesis, which results in a worse outcome in LIMD1 deficient patients, it is crucial to identify and target novel intrinsic vulnerabilities in these LIMD1 deficient lung cancers. Furthermore, this project also allows me to obtain my personal project licence to carry in vivo to validate the targets confirmed following the in vitro work.
Furthermore, a supplementary reason for me choosing this project was the lab group itself. In fact, being under the supervision of Prof. Sharp’s, as well learning from highly experienced postdoctoral researchers such as Dr. Kunal Shah and Dr. Paul Grevitt, were important aspects for me to consider for my optimal growth as a cancer researcher.
What skills have you developed so far? As I progressed through my MRes year and during my current initial period of my PhD, I have implemented my skills in key techniques such as routine cell culture, western blot, cell titer-glo and qPCR, to state a few. However, I am also fortunate to be learning and practising new fundamental methods in research like CRISPR technology. Increasing my knowledge in these essential research procedures and being able to master them is highly valuable for me to get the best quality of data during my PhD.
How have you found the mathematic/computational component of the DTP? The taught computational component of the DTP delivered at the University of Southampton represents a highly valuable opportunity for me to widen my skillset in terms of data analysis of large datasets. I am finding this module extremely interesting, and I believe it will be greatly useful for my academic career due to modern day cancer research requiring interdisciplinarity from different fields of study, especially bioinformatics and computational modelling.
What have you enjoyed most about your experience so far? What have you found challenging? So far, I am really enjoying my PhD as I am starting to organise, schedule and perform experiments after carefully planning for them. I highly enjoyed all three of my MRes rotations, as each of them provided me with a new skillset that I previously did not possess. As a full-time PhD student, carrying out experiments and maintaining an organised schedule is essential, but the laboratory environment can quickly evolve to a very fast-paced one with lots of challenges, for which I must be prepared to face.
 Elise Smith (Queen Mary)
Elise Smith (Queen Mary)
Cancer, Inflammation and Immunology
What rotation did you decide to pursue for your PhD project and why? I am undertaking my PhD in Dr Mirjana Efremova’s group, investigating the effect of chemotherapy on the single-cell landscape of colorectal cancer liver metastases. I chose this project as I am very interested in understanding how different cancer cell and tumour microenvironment populations are altered by therapy through non-genetic mechanisms. The opportunity to work with data from patient samples, and the major computational aspect of this PhD, were also key motivations for me.
What skills have you developed so far? In my current project, I have learnt and applied different methods for analysing single-cell RNA-seq data, including integration of datasets, differential expression and abundance testing, and inference of cell-cell communication. I will also be learning new computational methods for analysis of ATAC-seq and spatial transcriptomics data. During my MRes rotations, I developed wet-lab skills, such as immunohistochemistry and RNA-seq library preparation.
How have you found the mathematic/computational component of the DTP? The Quantitative Cell Biology module in Southampton was very helpful for learning how to analyse biological data in R, which I have applied for analysis of my own RNA-seq data. It was very valuable to discuss new bioinformatics concepts with researchers in the field and other students. The Queen Mary statistics module was also useful for understanding how to apply different statistical methods.
What have you enjoyed most about your experience so far? What have you found challenging? I have really enjoyed applying a number of different computational methods to explore patient-derived single-cell data in my project. I have enjoyed working collaboratively with other researchers and learning about the diverse areas of cancer research at the Barts Cancer Institute. It has sometimes been challenging to determine the best approach for analysing my data, in cases where there is not a readily-available or accepted method.
Ludovica Tarantola (Queen Mary)
Cancer, Inflammation and Immunology
 What rotation did you choose to pursue as your PhD project and why? My current PhD project with Dr Oliver Pearce aims to investigate the involvement of a particular extracellular matrix (ECM) protein, deposited by fibroblasts in the tumour microenvironment, in mediating resistance to chemotherapy in triple negative breast cancer (TNBC) via interactions with macrophages. I am going to characterise its regulation upon chemotherapy and immune checkpoint inhibitor (ICB) therapy in order to understand how to revert the immunosuppressive phenotype and to improve therapy response by using both in vitro and in vivo approaches.
What rotation did you choose to pursue as your PhD project and why? My current PhD project with Dr Oliver Pearce aims to investigate the involvement of a particular extracellular matrix (ECM) protein, deposited by fibroblasts in the tumour microenvironment, in mediating resistance to chemotherapy in triple negative breast cancer (TNBC) via interactions with macrophages. I am going to characterise its regulation upon chemotherapy and immune checkpoint inhibitor (ICB) therapy in order to understand how to revert the immunosuppressive phenotype and to improve therapy response by using both in vitro and in vivo approaches.
What skills have you developed so far? Thanks to the rotational year, I had the opportunity to widen and further develop several laboratory skills, such as western blotting, PCR, imaging techniques and analysis. Also, I have learnt cloning techniques and how to perform gene knockdown in vitro.
How have you found the mathematic/computational component of the DTP? It can be quite overwhelming at first, especially because this is something I have never been taught before. However, having an understanding of bioinformatics is becoming always more important within scientific research. Therefore, the quantitative biology module in Southampton is an important opportunity for learning basic R coding, which is an incredibly valuable skill to acquire for the future.
What have you enjoyed most about your experience so far? What have you found challenging? I have really enjoyed the rotational year I have just concluded. It was extremely useful to widen my research skills and to create a good network with other researchers in other departments working on different fields. I also really enjoy my current lab and department. It is an amazing environment to work in, I feel inspired everyday by the research of my colleagues and they are all really supportive and helpful. I have just started my PhD years, so I guess I still need to face the real challenges, even though the rotational year was also quite intense, with the exam workloads, lab experiments and research project write-up all at once!
Cohort 2020
 Jack Fisher (Southampton)
Jack Fisher (Southampton)
Immunity and Infection
What is your current project? I am investigating how a targeted cancer therapy used to treat B cell malignancies affects NK cell mediated immunity. As with many treatments, certain patients do not respond to this therapy, hence combination therapies are warranted. Targeted cancer therapies have been shown to modulate the NK cell immune response and given the promise of harnessing NK cells in immunotherapies, understanding how targeted cancer therapies modulate NK cell activity is key to optimising NK cell combination therapy approaches.
What skills have you developed so far, and what skills do you hope to develop? After one year I’ve learnt how to conduct complex flow cytometry experiments including NK cell activation and NK cell killing assays, perform western blots and tissue culture. In the future I am hoping to gain in vivo skills to study NK cell function in the tumour microenvironment and learn immunohistochemistry techniques to visualise NK cells in tissue. Alongside lab-specific skills, the programme has provided me with fundamental skills in scientific writing, enhanced my presentational capacity and improved my data handling in terms of storage, analysis and visualisation.
How have you found the mathematic/computational component of the DTP? It was a great opportunity to build upon the basic knowledge I obtained during my undergraduate studies. It was daunting at first, but after asking the incredibly helpful lecturers and student demonstrators (too) many questions it all clicked into place. I’ve not used what I learnt in my project yet, but so far it has been incredibly useful for understanding bioinformatic papers and when I do come round to performing bioinformatics I will certainly look back to this experience for guidance.
What have you enjoyed most about your experience so far? What has been challenging? I really enjoyed being able to rotate labs, one of the main reasons for choosing the programme. It enabled me to meet people from across the department to which I am new, but more importantly it allowed me to understand the speciality of each lab, therefore any scientific or technical problem I may encounter I have an array of people I can approach to help. I have enjoyed every experience so far, but the most challenging aspect of the course was juggling the 10-week lab rotation, writing the lab report and submitting assignments. These certainly kept me busy, but I wouldn’t change the experience one bit and the summer holiday definitely made up for those really busy periods of the year.
Mosammat Antara Labiba (Queen Mary)
Cancer, Inflammation and Immunology
What is your current project? I decided to pursue my first project with Dr Prabhakar Rajan and Professor Conrad Bessant as my PhD project. I will be using proteomics informed by transcriptomics (PIT) pipelines to identify putative neoantigens generated by alternative pre-mRNA splicing (AS). During AS, genes are shuffled to produce a number of protein isoforms from a single gene, which increases proteomic diversity. AS is disrupted in cancer, which may lead to new proteins that are processed as neoantigens for the cell. Tumours with a high neoantigen burden, typically arising from mutations, are effectively targeted by immune checkpoint-based anti-cancer immunotherapy. Hence, a greater understanding of the role of AS in neoantigen generation may reveal novel anti-cancer therapies.
I chose this project as it applies computation pipelines and artificial intelligence algorithms, which will equip me with the important skills necessary to advance in cancer research. Finally, having both a clinician scientist and computational biologist as my supervisors has ensured I get the best well-rounded support during my PhD.
What skills have you developed so far, and what skills do you hope to develop? I have developed important independence and organisational skills when planning my PhD research. The rotations also gave me a chance to gain wet-lab experience which I had not previously undertaken. I am also hoping to continue building on my coding skills in different languages to advance in my research.
How have you found the mathematic/computational component of the DTP? I had previously completed an MRes in Cancer Informatics at Imperial College London, which equipped me with the necessary R skills needed for biological research. Therefore, the bioinformatics/computational component of the programme was not as demanding for me.
What have you enjoyed most about your experience so far? What have you found challenging? I enjoyed the chance to undertake three different projects before choosing my final PhD project. I got the chance to try out new experiences for example in wet-lab which I may not have had I gone directly into a PhD. I think this opportunity to try out a lab before committing to the full PhD is indispensable and I am extremely grateful for this opportunity. I also really enjoyed my optional module “Infection and Immunity” which was taught at Southampton (due to Covid it was online) alongside my second module.
I least enjoyed the two taught modules during the first semester as I was unable to focus enough time on my rotation project in comparison to the other two projects.
Mara Gelmetti (Queen Mary)
Cancer, Inflammation and Immunology
 What is your current project? I am currently undertaking my PhD in Dr. Inês Sequeira’s lab. My PhD project looks at understanding the role Keratin 76 plays on the immune system. Previous studies from the lab reported an immunomodulatory role for Keratin76 during carcinogenesis in mice, my project aims to understand the mechanism driving this phenotype. I was attracted by the multidisciplinary nature of the project which combines aspects of immunology, cell biology and developmental biology.
What is your current project? I am currently undertaking my PhD in Dr. Inês Sequeira’s lab. My PhD project looks at understanding the role Keratin 76 plays on the immune system. Previous studies from the lab reported an immunomodulatory role for Keratin76 during carcinogenesis in mice, my project aims to understand the mechanism driving this phenotype. I was attracted by the multidisciplinary nature of the project which combines aspects of immunology, cell biology and developmental biology.
What skills have you developed so far, and what skills do you hope to develop? Thanks to the multidisciplinary aspect of the project, I have been fortunate enough to develop a wide range of laboratory skills. These include flow cytometry, immunostainings and some molecular biology techniques such as PCR. As the project develops, I hope to develop more bioinformatic skills to analyse scRNA-seq data.
How have you found the mathematic/computational component of the DTP? The bioinformatics modules were incredibly helpful. I am very grateful that we were given dedicated time and support in understanding bioinformatics concepts and then applying these to real data sets. Although challenging throughout, at the end of the course I really felt like I had a strong foundation to help me approach future analyses in R.
What have you enjoyed most about your experience so far? What have you found challenging? I really enjoyed the rotational aspect of the programme, and the opportunities to work with different teams. The infection and immunity module was also incredibly helpful as it was structured in the form of a weekly journal club. Although challenging to time-manage a research project and coursework, it was a great exercise for a scientific career.
Alba Gonzalez (Southampton)
Centre for Cancer Immunology
What is your current project? My project aims to understand how monoclonal antibodies can boost the immune system to target tumours whilst minimising toxic effects. In particular, I am investigating the therapeutic efficacy of several monoclonal antibodies against a receptor called 4-1BB. This receptor is an interesting target because it is expressed on CD8+ T cells and can promote their cytotoxic potential and survival, improving their anti-tumour activity.
What skills have you developed so far, and what skills do you hope to develop? I have learnt a range of lab techniques such as flow cytometry, Surface Plasmon Resonance and molecular biology techniques including PCR. Also, during my second rotation I got to experience techniques involved in protein and antibody production, such as cell transfections and protein purification. I hope to further develop my molecular biology and cloning skills, as well as to using in vivo models to test the therapeutic efficacy of different antibodies.
How have you found the mathematic/computational component of the DTP? The Quantitative Cell Biology module has been very valuable to help me to gain an insight into large datasets. Also, it was great to get an overview of how to analyse genomics, transcriptomics and proteomics data. Although my PhD project is lab-based, knowing about bioinformatics has been really helpful to understand papers, seminars and talks!
What have you enjoyed most about your experience so far? What have you found challenging? The best part of the iPhD programme is experiencing different labs during the rotation year. It’s a great opportunity to sample the projects but also to meet people in different labs and to learn different techniques that you can incorporate into your final PhD project. Also, being part of a cohort of 7 students was incredibly helpful to settle into the programme, especially during a global pandemic!
Rosa Alves Martins (Southampton)
Centre for Cancer Immunology
 What is your current project? I am currently working on identifying targetable proteins on the surface of tumour-associated macrophages (TAMs) that infiltrate a low-oxygen tumour microenvironment. Hypoxia can influence the behaviour of TAMs by promoting their pro-tumour role, leading to increased therapy resistance. This research will help improve our understanding of the effect hypoxia has on TAMs, how to identify them, target them and subsequently find ways to improve treatment efficacies.
What is your current project? I am currently working on identifying targetable proteins on the surface of tumour-associated macrophages (TAMs) that infiltrate a low-oxygen tumour microenvironment. Hypoxia can influence the behaviour of TAMs by promoting their pro-tumour role, leading to increased therapy resistance. This research will help improve our understanding of the effect hypoxia has on TAMs, how to identify them, target them and subsequently find ways to improve treatment efficacies.
What skills have you developed so far, and what skills do you hope to develop? I have developed a wide range of both wet and dry laboratory skills from completing three distinct rotation projects. For example, I learnt about flow cytometry, I familiarised myself with analysing transcriptome data with R Studio and I learnt how to process human leukocytes. Additionally, this programme has allowed me to improve my scientific writing skills and expand my knowledge of immunology. As the project progresses, I aim to develop skills in tissue staining, cell sorting, and spatial transcriptomics to better understand the hypoxic tumour microenvironment.
How have you found the mathematic/computational component of the DTP? I found the bioinformatics component of the programme to be beneficial, although it was also quite overwhelming at the beginning due to being a novice. Also, the programme leads were always available to help and with practice, it did become more intuitive. Indeed, I believe it is important for scientists nowadays to have some knowledge and understanding of bioinformatics and the role it plays in scientific research.
What have you enjoyed most about your experience so far? What have you found challenging? I enjoyed the format of the rotation projects, getting to learn new techniques and gain a greater understanding of cancer immunology. It also helped me to become more confident in the lab as well as more independent. Additionally, because more people have done the programme previously, it meant I could ask for advice and support from students in the previous cohort, who are always more than happy to help. As with all experiences, there are some drawbacks, namely, I found the constant workload with few breaks overwhelming at times. However, I did find that the challenge allowed me to improve my ability to work under pressure and hone my time-management skills.
Cohort 2019
 Amanda Stafford (Queen Mary)
Amanda Stafford (Queen Mary)
Cancer Inflammation and Immunology
What is your current rotation? I am currently doing a rotation in Stephanie Kermorgant’s lab, investigating whether the cMet signalling pathway, stimulated by HGF, leads to an increase in PDL1 expression.
What skills have you developed so far, and what skills do you hope to develop? I have learnt how to perform western blotting. I think I will go on to develop skills in immunofluorescence and microscopy.
How have you found the mathematic/computational component of the DTP? It's been incredibly valuable to have assigned time to learn how to use R. It's something I think will be incredibly useful in my research career, and I just don’t think I could have invested the time independently to learn it.
What have you enjoyed most and least about your experience so far? I've really enjoyed joining a PhD scheme with three other people. I think the taught computational module has been the most useful. I have least enjoyed the research methods module; it, and its coursework, are quite time consuming.
Kamila Bienkowska (Southampton)
Cancer Immunology

What is your current rotation? I am currently undertaking a research project at Prof. Gareth Thomas' lab under the supervision of Dr Christopher Hanley. I analyse single cell RNA sequencing data from cancer patients and investigate the heterogeneity of cancer-associated fibroblasts (CAFs) found within the tumours. CAFs are components of the tumour microenvironment known to aid cancer progression. Understanding the mechanism through which they act could improve future cancer therapies.
What skills have you developed so far, and what skills do you hope to develop? I have learnt a few new research methods that I hadn't yet had the chance to perform, including flow cytometry and immunoprecipitation. I picked up bioinformatics during the rotations and I keep working on these new skills. Also, as a student for whom English is not their first language, I got to work on my writing skills which I think will be very useful in the future. As for the skills that I'd like to develop, I would enjoy the opportunity to work with, amongst others, immune cells and with primary cells.
How have you found the mathematic/computational component of the DTP? It felt a little overwhelming at first, but from the time perspective I am confident to say that it was very helpful in learning the basics of R coding. Nowadays, bioinformatics analysis is a desirable skill and so being introduced to it at the beginning of the PhD was important. Continuing my bioinformatics adventure during the rotation projects made me want to incorporate it in my final PhD project, despite being terrified of it at first.
What have you enjoyed most and least about your experience so far? The best part of the programme is being able to test three different potential projects and choose the one that you like the most. The first three months of the programme were the busiest with assignments, so I enjoyed it the least, but I guess that's a "student's life."
Matthew Beck (Queen Mary)
Cancer Inflammation and Immunology
 What rotations have you completed so far? My first rotation was under the supervision of Dr Pradeep Madapura at the Blizard Institute and was titled "Generation of MSL and MOF Knockdown Cell Lines to Study Their Role in Enhancer Function and Gene Regulation." In this project I was trying to produce stable gene knockdown cell lines in which the histone acetyltransferase proteins MSL and MOF could be studied in order to shed light on their role in development and cancer.
What rotations have you completed so far? My first rotation was under the supervision of Dr Pradeep Madapura at the Blizard Institute and was titled "Generation of MSL and MOF Knockdown Cell Lines to Study Their Role in Enhancer Function and Gene Regulation." In this project I was trying to produce stable gene knockdown cell lines in which the histone acetyltransferase proteins MSL and MOF could be studied in order to shed light on their role in development and cancer.
I undertook my second rotation in Dr Gunnel Hallden’s lab at Barts Cancer Institute, working on oncolytic adenoviral therapy for the treatment of prostate cancer. The aim of this project was to develop new methods, such as combining with peptides, to enhance the delivery of the therapeutic adenoviruses to the prostate cancer cell and ultimately increase cell killing.
What skills have you developed so far, and what skills do you hope to develop? I have been fortunate to develop a wide range of laboratory skills over the first two rotations. These include a number of fundamental research techniques such as cell culture, PCR, western blotting just to name a few, and I feel that this will stand me in strong stead for work that I will encounter throughout and beyond my PhD. As my third rotation will be in the form of a literature review, I really hope to be able to develop my ability to critically review other research papers.
How have you found the mathematic/computational component of the DTP? One hugely interesting aspect of this course is the taught computational module that is undertaken in Southampton. This was a real eye opener for me as I had previously no experience in this at all and went into the course fairly nervous about it. It was so encouraging that by the end of the module I felt I had a solid foundation understanding of bioinformatics and how it can be applied. I look forward to trying to develop this more during my PhD.
What have you enjoyed most about your experience so far? I think the most enjoyable aspect of the programme is without doubt the opportunity to spend time in different labs doing completely different types of research. Not only do you learn a wide range of different skills and see how different researchers work, but it allows you to make a very informed decision on which project you wish to select for the PhD.
Steven Turner (Southampton)
Cancer Immunology
 What is your current rotation? I am studying regulation of the immune response in relation to peptide immunogenicity - very important concepts in understanding targeting of cancer cells and viruses. I am currently working to investigate the conformational dynamics of MHC-I proteins by combining experimental hydrogen-deuterium exchange (HDX) datasets and computational molecular dynamics simulations. HDX is limited as deuterium uptake is measured across entire peptide fragments, preventing insight at the single residue level. By combining the high-resolution conformations sampled by MHC proteins during molecular dynamics simulations we can predict the probable structures present in the HDX experiments. This can help provide insight into how different MHC alleles are able to select for certain peptides and vary their dependency on chaperones such as tapasin and ERp57.
What is your current rotation? I am studying regulation of the immune response in relation to peptide immunogenicity - very important concepts in understanding targeting of cancer cells and viruses. I am currently working to investigate the conformational dynamics of MHC-I proteins by combining experimental hydrogen-deuterium exchange (HDX) datasets and computational molecular dynamics simulations. HDX is limited as deuterium uptake is measured across entire peptide fragments, preventing insight at the single residue level. By combining the high-resolution conformations sampled by MHC proteins during molecular dynamics simulations we can predict the probable structures present in the HDX experiments. This can help provide insight into how different MHC alleles are able to select for certain peptides and vary their dependency on chaperones such as tapasin and ERp57.
What skills have you developed so far, and what skills do you hope to develop? I have continued developing my lab-based skills in western blotting, FACS, tissue-culture and more. I have also learnt completely new skills in molecular dynamics simulations and the associated analysis of protein trajectories. I hope to learn to apply advanced sampling methods to my MD simulations as well as conduct my own HDX experiments in the future.
How have you found the mathematic/computational component of the DTP? An introduction to basic R coding skills and getting to apply this to biological datasets is a useful foundation to modern data analysis that is becoming an increasingly more critical skill for research scientists to have. Consequently, without a strong grasp of the underlying statistics these methods can often be easily misrepresented, which is why I believe combining these two subjects into this course has been an invaluable addition to my skillset as an early career researcher.
What have you enjoyed most about your experience so far? The opportunity to tailor my projects to what I believed benefited me most has put me in a position where I feel very prepared for my future PhD project. I have been able to gain experience in a wide array of subjects from lab-based biochemistry to pure computational chemistry, all of which have provided unique skills I plan to utilise in the future.
Emily O'Sullivan (Queen Mary)
Cancer Inflammation and Immunology
 What is your current rotation? The title of my current rotation project is 'Investigating the impact of eradicating cancer cells with amplified centrosomes in cancer.' Centrosome amplification is a hallmark of cancer and has been shown to promote tumorigenesis in vivo, leading to chromosome instability (CIN) and increased invasive capacity of cells. I am currently helping to generate a new cell line to investigate what happens to the tumour microenvironment (TME) when cells with extra centrosomes are removed.
What is your current rotation? The title of my current rotation project is 'Investigating the impact of eradicating cancer cells with amplified centrosomes in cancer.' Centrosome amplification is a hallmark of cancer and has been shown to promote tumorigenesis in vivo, leading to chromosome instability (CIN) and increased invasive capacity of cells. I am currently helping to generate a new cell line to investigate what happens to the tumour microenvironment (TME) when cells with extra centrosomes are removed.
What skills have you developed so far, and what skills do you hope to develop? So far I have developed both my cellular and molecular biology skills through experience with gateway cloning and immunofluorescence staining. I have also become far more confident with microscopy and tissue culture. Moving forward in the project I hope to develop skills in tissue staining and identifying TME components.
What have you enjoyed most and least about your experience so far? I am thoroughly enjoying becoming confident working independently in the lab, developing my own ideas and expanding my current knowledge of the cell cytoskeleton. It has been a challenge to become accustomed to managing my own time and not allowing the work load to become overwhelming.
Timo Kuerten (Southampton)
Centre for Cancer Immunology
 What is your PhD project focused on? My PhD project, in collaboration with Swedish biotechnology company BioInvent, is aimed at finding novel tumour T-regulatory cell (Treg) therapeutic targets and then developing antibodies against them. Tregs are a sub-population of CD4 T cells that suppress the body’s immune response against the tumour, so depleting them from the tumour microenvironment (TME) increases the effectiveness of existing immunotherapies that expand cytotoxic CD8 T cells, leading to improved tumour control in more patients. In terms of methodology, my project uses a blend of dry and wet lab techniques, where I’ve created a bioinformatic pipeline to identify new potential Treg targets, and I am now back in the lab to characterise the targets I’ve identified and generate antibodies against them.
What is your PhD project focused on? My PhD project, in collaboration with Swedish biotechnology company BioInvent, is aimed at finding novel tumour T-regulatory cell (Treg) therapeutic targets and then developing antibodies against them. Tregs are a sub-population of CD4 T cells that suppress the body’s immune response against the tumour, so depleting them from the tumour microenvironment (TME) increases the effectiveness of existing immunotherapies that expand cytotoxic CD8 T cells, leading to improved tumour control in more patients. In terms of methodology, my project uses a blend of dry and wet lab techniques, where I’ve created a bioinformatic pipeline to identify new potential Treg targets, and I am now back in the lab to characterise the targets I’ve identified and generate antibodies against them.
What skills have you developed so far? I have learnt a wide variety of skills since starting this programme, whether that be wet lab, dry lab or transferrable skills. Wet lab skills that I have acquired or developed further include classical skills such as qPCR, molecular biology, western blots, fluorescence microscopy and flow cytometry, but also more specialised skills such as organoid growth and manipulation as well as time-lapse microscopy. I have also picked up brand new computational skills, specifically learning how to use the R programming language to perform transcriptomic analyses. In terms of transferrable skills, this programme has allowed me to develop my scientific writing skills through writing up a report for each rotation, and heightened my critical analysis skills through the critical examination of published high-impact papers. I have also been able to develop my immunological knowledge in more depth. Finally, collaborating closely with BioInvent has allowed me to experience the industry side of cancer research, which I am hoping to learn even more about on my trip to Sweden to work in their labs!
What have you enjoyed most and least about your experience so far? I believe one of the most valuable aspects of the integrated PhD programme is getting the chance to work on three different projects with three different PIs, and then choose the one that’s the best fit for you. This means that you’re almost guaranteed to end up working on something you enjoy, with a supervisory style that suits your own working/learning pattern. With the iPhD programme there is even the scope to design your own project if that’s up your alley! Personally, I’ve also really benefitted from the individual attention invested in each student, which really makes you feel like a valued member of the department. As with all things, there are ups and there are some downs. This is an intense programme, and the workloads and deadlines are not few. It can sometimes be hard to manage full-time lab work whilst also finding additional hours in the week to work on other deadlines such as conferences and progression reviews; but at least this is a good way to continue learning how to work under pressure and improve project-management skills in the process.
Molly Guscott (Queen Mary)
Cancer Inflammation and Immunology
 What is your current rotation? The title of my current rotation project is 'Discovering the role of PARP1 BRCT domain in DNA repair and elucidating the potential of it's inhibition in cancer treatment.' This rotation project with Zuzana Horesji was aimed at the understanding of DNA damage response proteins. We are looking at HRPU and other HRPU homology proteins, in particular their expression levels at the protein level in response to damage, their activity before and after phosphorylation and how this affects function. This is using cloning techniques to produce novel truncation of the protein of interest as well as using cell culture followed by immunoprecipitation to investigate protein interactions.
What is your current rotation? The title of my current rotation project is 'Discovering the role of PARP1 BRCT domain in DNA repair and elucidating the potential of it's inhibition in cancer treatment.' This rotation project with Zuzana Horesji was aimed at the understanding of DNA damage response proteins. We are looking at HRPU and other HRPU homology proteins, in particular their expression levels at the protein level in response to damage, their activity before and after phosphorylation and how this affects function. This is using cloning techniques to produce novel truncation of the protein of interest as well as using cell culture followed by immunoprecipitation to investigate protein interactions.
What skills have you developed so far? So far, skills developed have been predominantly in the laboratory environment, particularly cloning techniques. As well as that, the Southampton computational module has been quite enlightening and I have come out with a working understanding of R coding as well as it uses for 'omics research.
How have you found the mathematic/computational component of the DTP? The bioinformatics component taught at Southampton is structured in a very good way and helps initially with getting to grips with R coding. However after this initial session with R the level of coding increases exponentially. Looking at single cell sequencing, principal component analysis etc is quite difficult to understand. The coursework for this section is quite helpful to get to grips with the techniques.
What have you enjoyed most and least about your experience so far? The most enjoyable aspect of the course for me so far has been learning to use R to a standard which is actually useful. The least enjoyable aspect is balancing lab work with lectures and coursework.
Cohort 2018
 Nicolas Roth (Queen Mary)
Nicolas Roth (Queen Mary)
Cancer Inflammation and Immunology
Which rotation did you decide to pursue for your PhD project? I decided to continue with my third rotation for my PhD project, which looked at the role of HER2-CB2 heteromers in the progression of breast cancer. HER2 is an established prognostic marker for invasive breast cancers and HER2 expression in breast cancer can identify patients at risk of progressing to the invasive disease. The cannabinoid receptor 2 (CB2) is closely correlated with HER2 expression in breast cancer patients and administration of Δ9-tetrahydrocannbinol (THC) leads to the senescence of cancer cell growth and an increase in apoptosis. THC was found to cause the dissociation of the HER2-CB2R heteromer and lead to proteasomal degradation of HER2, preventing tumourogenic signalling. In this rotation I developed a 3D model of breast cancer using the breast cell line MCF10A, with inducible expression of HER2 and CB2, to investigate the functional role of these receptors in breast cancer progression. I decided to continue with this rotation for my PhD project due to the importance of 3D models in in vitro biology, the level of collaboration this project involves and the fascinating biology. I will continue to study the role of HER2 and CB2 interaction in breast cancer and how these receptors influence signalling, metabolism and the overall progression of breast cancer.
What skills have you developed so far? The rotation year offered valuable experience developing a wide range of lab skills and the ability to plan and carry out experiments independently. Throughout my PhD I expect to be competent at many techniques in the lab, whilst also developing my data analysis and interpretation, presentation skills and critical analysis of other scientists’ work.
Have you contributed to any publications so far? I currently am a co-author on a paper that is under review.
Have you attended, presented a poster or spoken at any conferences? I attended the Biochemical Society conference: Cell Signaling and Intracellular Trafficking in Cancer Biology: Interplay, Targeting and Therapy in Turin Italy on 21-24 October 2019.
Laura Reid (Southampton)
Immunity and Infection Pathway
 What rotations did you complete during your first year of the programme? I completed the following rotations: Production of a recombinant fragment of human surfactant protein A (SP-A) using a mammalian expression system; Petri net computational modelling of dendritic cell gene regulatory network in response to Plasmodium falciparum; and The cellular origin of microRNA dysregulated in chronic obstructive pulmonary disease (COPD) lung-derived extracellular vesicles (EVs).
What rotations did you complete during your first year of the programme? I completed the following rotations: Production of a recombinant fragment of human surfactant protein A (SP-A) using a mammalian expression system; Petri net computational modelling of dendritic cell gene regulatory network in response to Plasmodium falciparum; and The cellular origin of microRNA dysregulated in chronic obstructive pulmonary disease (COPD) lung-derived extracellular vesicles (EVs).
What did you choose as your PhD project and why did you choose it? My PhD project investigates the hypothesis that microRNA differentially expressed in COPD lung-derived EVs compared to healthy controls are selectively packaged and released from epithelial cells as a form of intercellular communication mediating chronic inflammation in COPD. I chose this project after being introduced to the field of EVs in my final MRes rotation project with the Pulmonary Immunology Group. Over recent years the role of EVs in disease has become an exciting and quickly evolving area of research with many novel diagnostic and therapeutic applications being investigated. In addition I chose to join the Pulmonary Immunology Group as they offered the knowledge and support required to complete a successful PhD project.
What skills have you developed so far? I have so far developed a wide range of technical and transferable skills relevant to my development as a professional researcher. During the MRes I gained valuable skills in statistical and bioinformatic analyses from the taught modules and rotation projects which are really useful both in terms of my current PhD project and future career. I have also gained experience in a variety of laboratory techniques including tissue culture, recombinant protein synthesis, western blot analyses and quantification of microRNAs using real-time PCR. Furthermore I developed skills including communicating with outreach and critical analyses of research. Over the next few years I wish to develop skills including microscopy and making an impact with my research.
Have you attended, presented a poster or spoken at any conferences? I presented at the IPhD and MRC DTP AwayDay in Southampton (2019) and attended the UK Extracellular Vesicles Forum (2019).
Have you completed any additional training? I have completed the Introductory course to 2D image processing and analysis.
What have you enjoyed about your experience so far? Overall what I have enjoyed so far about the iPhD programme is the support and opportunities available to allow you to identify and pursue an area of research you are passionate about.
Robert Hearnden (Queen Mary)
Cancer Inflammation and Immunology
 Which rotation did you decide to pursue for your PhD project? My PhD project is entitled 'Investigating cell intrinsic metabolism of dendritic cells' and is a continuation of my first rotation. I chose this project for a number of reasons. Firstly, it was the project which I was most passionate about. Also, I felt that this project has a strong potential to be fully translatable, from the basic biology, right through to validation in human tissue and identification of potential drug targets for use in clinics. In my particular case I hope to eventually apply my research in a breast cancer setting. Another reason for selecting this project was my supervisors. Between them, they have strong expertise in the fields of immunology and metabolism, with my primary supervisor also being in charge of the metabolomics core facility at Barts Cancer Institute.
Which rotation did you decide to pursue for your PhD project? My PhD project is entitled 'Investigating cell intrinsic metabolism of dendritic cells' and is a continuation of my first rotation. I chose this project for a number of reasons. Firstly, it was the project which I was most passionate about. Also, I felt that this project has a strong potential to be fully translatable, from the basic biology, right through to validation in human tissue and identification of potential drug targets for use in clinics. In my particular case I hope to eventually apply my research in a breast cancer setting. Another reason for selecting this project was my supervisors. Between them, they have strong expertise in the fields of immunology and metabolism, with my primary supervisor also being in charge of the metabolomics core facility at Barts Cancer Institute.
What skills have you developed so far? So far during my rotations I have improved my abilities to communicate my research. I have gained experience in a number of techniques: immunohistochemistry, western blot, immunoprecipitation, primary cell culture, microscopy, flow cytometry, handling RNA sequencing data, qPCR. I have also developed my time management and organisation skills.
What skills do you think you will develop going forward? In the future I will gain technical experience in running samples on an orbi-trap mass spectrometer. I will further improve my analytical and experimental design skills. I will also gain skills in more advanced cell culture techniques, and metabolic profiling of cells. Lastly, I will be handling big data sets and developing my ability to extract useful information from these data sets.
How did you find the mathematics/computational component of the DTP? The bioinformatics component of the DTP was very useful, now I feel like, with a little bit of time, I’d manage to work through most data sets. It also gave me the confidence to proactively seek opportunities to use these skills in my rotation projects, and I hope eventually in my PhD project.
Have you completed any additional training so far? I have completed the Home office licensee training course (PILA-C) for my personal license.
What have you enjoyed most and least about your experience so far? I have enjoyed most the opportunity to choose whatever three rotation projects I would like, and as a result, I now have vastly increased my network of scientists within the School of Medicine and Dentistry. I least enjoyed juggling my first rotation with one day a week in Southampton and the taught component on a Wednesday.
Gabriela Virdzekova (Southampton)
Cancer Sciences Pathway
 About me: Before commencing the PhD, I have obtained my undergraduate degree in Biomedical Science at the University of Southampton and undertook an internship at the German Cancer Research Center. During my studies, I really enjoyed immunology and cancer biology and decided to pursue a PhD at the University of Southampton, which is at the forefront of cancer immunology and immunotherapy research.
About me: Before commencing the PhD, I have obtained my undergraduate degree in Biomedical Science at the University of Southampton and undertook an internship at the German Cancer Research Center. During my studies, I really enjoyed immunology and cancer biology and decided to pursue a PhD at the University of Southampton, which is at the forefront of cancer immunology and immunotherapy research.
What is your PhD project? My project is studying T cells and how they kill cancer cells. I am looking at the role of Akt signalling pathway in CD8+ T cells. We are investigating whether modulation of this signalling pathway could be used to improve response to immune checkpoint blockers during cancer therapy. I am currently focusing on determining whether inhibition of Akt pathway changes CD8+ T cell ability to kill cancer cells and facilitate immune response in the tumour microenvironment.
Which skills have you developed so far? Through my PhD, I have developed a variety of lab skills, including western blotting, cell culture, flow cytometry, qPCR, polysome profiling and in vivo work. I have also developed skills in bioinformatics during the analysis of single-cell RNA sequencing data. I aim to further expand my skills by analysing bulk RNA sequencing data during the remainder of my PhD. I have gained organisational and time management skills, writing skills and have become more confident during public speaking.
Have you completed any additional training? I have completed the Home Office licensee training course necessary for my research.
What have you enjoyed most and least about your experience so far? I really enjoy working in a sociable and supportive environment and being surrounded by highly ambitious academics and students. Learning bioinformatics was a very enjoyable experience and I am planning to continue developing this skill in the future. I anticipate there will be more opportunities to do another internship to develop skills outside our department.
Reza Roozitalab (Queen Mary)
Cancer Inflammation and Immunology
 What is your PhD project and how did you choose it? My PhD project is to investigate the functional relationship of macrophages in DCIS progression with a focus on the fate of myoepithelial cells. Using and developing a 3D in vitro model of ductal carcinoma in situ (DCIS). My aim is to assess the behaviour of myoepithelial cells in response to primary macrophages of different phenotypes. Characterising DCIS associated macrophages will form an important part of my project. I chose this project due to its clinical relevance and focus on 3D in vitro modelling.
What is your PhD project and how did you choose it? My PhD project is to investigate the functional relationship of macrophages in DCIS progression with a focus on the fate of myoepithelial cells. Using and developing a 3D in vitro model of ductal carcinoma in situ (DCIS). My aim is to assess the behaviour of myoepithelial cells in response to primary macrophages of different phenotypes. Characterising DCIS associated macrophages will form an important part of my project. I chose this project due to its clinical relevance and focus on 3D in vitro modelling.
Which skills have you developed so far? I have developed a great deal of independence both at the bench and away developing and planning ideas. The three short rotation projects have taught me that, while things may not always go to plan, there is always a lesson to take away for next time.
Which skills do you think you will develop going forward? I think I will continue to become more independent in making plans and decisions that will shape my final PhD thesis and understand the commitment that is required to be in academia and lead a research group.
How did you find the mathematics/computational component of the DTP? The mathematics/computations component was a full on introduction to R with real datasets to manipulate and understand. While daunting at first for a complete beginner, it has opened my eyes to the value of computational biology in complementing our traditional research.
What have you enjoyed most and least about your experience so far? What I have enjoyed most is being able to work on three short projects with an opportunity to tailor each in line with my specific interest. I would have enjoyed committing 100% of my time to my lab project rather than coursework etc.
Ottilie Swinyard (Queen Mary)
Cancer Inflammation and Immunology
 What is your PhD project and how did you choose it? I chose to continue with my second rotation with Professor Trevor Graham as my PhD project, which I'm now carrying out on the early immune and genetic events in Lynch syndrome patients and the progression to colorectal cancer. I decided to choose this project due to learning the most while performing my rotation with this group, as well as having a particular interest in the interactions between genomic alterations and the microenvironment contributing to the initiation of cancer. This group is very diverse, containing computational mathematicians, clinical fellows, immunologists and wet lab researchers which makes it a dynamic and inspirational environment to be in.
What is your PhD project and how did you choose it? I chose to continue with my second rotation with Professor Trevor Graham as my PhD project, which I'm now carrying out on the early immune and genetic events in Lynch syndrome patients and the progression to colorectal cancer. I decided to choose this project due to learning the most while performing my rotation with this group, as well as having a particular interest in the interactions between genomic alterations and the microenvironment contributing to the initiation of cancer. This group is very diverse, containing computational mathematicians, clinical fellows, immunologists and wet lab researchers which makes it a dynamic and inspirational environment to be in.
Which skills have you developed so far? So far during my MRC studentship I have learnt a lot of computational analysis from scratch, such as coding, statistics and AI image analysis. Along with laboratory techniques such as siRNA, CRISPR, RNA/RNA extraction and microscopy techniques.
Which skills do you think you will develop going forward? I'd like to learn single cell techniques like single cell RNA sequencing and further my computational analysis skills.
How did you find the mathematics/computational component of the DTP? The taught MRC computational module gives a very good grounding in the most common types of analysis carried out and basic R coding, which can then go on to be used and developed within the rotations performed.
Have you contributed to any publications so far? I am currently working on a review.
What have you enjoyed most and least about your experience so far? I really enjoyed my three rotations in first year. I think this is an excellent opportunity to get to know what you like best and least in the laboratory and also experience many different ways of working within a lab, ways Principal Investigators work differently and various fields within cancer and immunology.
Shaun Maguire (Southampton)
Centre for Cancer Immunology
 What is your PhD project and why did you choose it? My project is 'Investigating the therapeutic potential of aGITR (glucocorticoid-induced TNF receptor-related gene) antibodies.' GITR is a co-stimulatory receptor found on the surface of immune cells. In this project, I am testing the therapeutic efficacy of anti-aGITR antibodies produced by the University of Southampton Antibody and Vaccine group, for immunomodulatory tumour therapy. I chose this project because developing therapeutic antibodies is an exciting topic and the university has many research groups that are experts in this field.
What is your PhD project and why did you choose it? My project is 'Investigating the therapeutic potential of aGITR (glucocorticoid-induced TNF receptor-related gene) antibodies.' GITR is a co-stimulatory receptor found on the surface of immune cells. In this project, I am testing the therapeutic efficacy of anti-aGITR antibodies produced by the University of Southampton Antibody and Vaccine group, for immunomodulatory tumour therapy. I chose this project because developing therapeutic antibodies is an exciting topic and the university has many research groups that are experts in this field.
What skills have you developed so far? In the lab, I have developed skills in flow cytometry, tissue culture, Biacore, 3D culture of organoids, DropSeq, western blots, analysis of single cell RNA seq datasets and IHC. Other skills I have developed include thesis writing, presenting and the critical appraisal of research articles. I hope to continue to develop my writing skills, presenting skills and in vivo animal work.
How did you find the mathematics/computational component of the DTP? Both RPB and RPC involved analysis of scRNA seq datasets which I really enjoyed. It was nice to develop these skills as computational methods are becoming more prevalent in scientific research.
How have you found your experience so far? There was a really interesting variety of projects available as part of my rotation year. Because of this, I have been able to develop a variety of different skills. The support and guidance throughout has been excellent. I’m happy with how things are going and enjoying the research I am a part of. The away day in my first year was interesting, I got to see the scope of research being undertaken at both the University of Southampton and Queen Mary University of London.
George Elder (Queen Mary)
Cancer Inflammation and Immunology
What is your PhD project and how did you choose it? The diverse and highly complex nature of modern biological research produces a high volume of data. Thus, there is unprecedented need to develop tools and methodologies to explain and rationalise these results. The aim of my PhD project is to develop novel logic-based algorithms that overcome the limitations of existing tools used for analysis. Ultimately, the most challenging aspect of this work will be to develop an automated hypotheses generation and validation algorithm. This will be able to employ abductive reasoning in combination with scientific knowledge and expertise to logically analyse not only datasets which suffer from the issues mentioned above but others as well. The resulting hypotheses can then be validated entirely in silico based on information garnered from other databases. If this yields interesting and novel results these can ultimately be tested by carrying out the relevant physical experiments manually.
What skills have you developed so far? I have expanded my skills on general computer-based analysis. Namely, following various courses I have attended at Queen Mary, I have gained more advanced skills in Python-based data analysis. As the main focus of my project is applying a novel approach via logic programming, I have gained a substantial working background, which I am continuously expanding, on the use and application of the LP language Prolog, specifically its SWI-Prolog implementation.
Have you attended, presented a poster or spoken at any conferences? I attended two conferences in 2019. The first was the American Society for Mass Spectrometry (ASMS) conference in Atlanta, Georgia. Here I presented a poster detailing the initial stages of my work relating to explaining the phosphoproteomics dataset I am working on. The second conference I attended was the International Conference on Logic Programming (ICLP) at Las Cruces, New Mexico. As part of the conference I attended a 2-day intense course titled 'Autumn School on Logic Programming' as well as a 1-day workshop on Probabilistic Logic Programming. Through these, apart from a plethora of ideas to pursue as part of my own work, I made a number of connections with potential collaborators.
Have you completed any additional training? I have been accepted as part of the PhD Enrichment scheme at the Alan Turing Institute. For this I will be based at the Institute for six months beginning January of 2020. Here, I will have the chance to collaborate with the researchers within the Institute as well as the various Fellows that are part of it but based elsewhere.
Cohort 2017
Atiya Sarmin (Queen Mary)
Cancer Inflammation and Immunology
 About me: I received my BSc Biomedical Sciences degree from the University of Southampton where I first became interested in Cancer Immunology. This led me to complete an MRes in Cancer Biology at Imperial College London, where I worked in Ovarian Cancer Research and Glioblastoma Research labs. I then joined this MRC DTP at Barts and the London School of Medicine and Dentistry.
About me: I received my BSc Biomedical Sciences degree from the University of Southampton where I first became interested in Cancer Immunology. This led me to complete an MRes in Cancer Biology at Imperial College London, where I worked in Ovarian Cancer Research and Glioblastoma Research labs. I then joined this MRC DTP at Barts and the London School of Medicine and Dentistry.
Have you attended, presented a poster or spoken at any conferences? I gave an oral presentation at Termis-EU 3D Bioprinting in Cancer Research (2019). I also gave an oral presentation at the International 3D Bioprinting Research Symposium (2019), where I was winner of the young investigator’s oral presentation competition. I gave a poster presentation at BioMedEng (2019).
What have you enjoyed the most and the least about your experience so far? I have mostly enjoyed learning a whole new area of research – bioinformatics. It has been very challenging, but it has been great to be able to use it in real research. It is hard to keep up with the workload as it is a very fast-paced programme, but it has been a new challenge and an enjoyable experience.
Charys Papagregoriou (Southampton)
Centre for Cancer Immunology
 What is your PhD project? My PhD project investigates the characterisation of LILRB3, a novel myeloid inhibitory receptor and the generation of a CAR T-cell immunotherapy against this receptor to treat acute myeloid leukaemia.
What is your PhD project? My PhD project investigates the characterisation of LILRB3, a novel myeloid inhibitory receptor and the generation of a CAR T-cell immunotherapy against this receptor to treat acute myeloid leukaemia.
What skills have you learnt so far? Since my project involves two separate aspects, I have gained a broad range of skills to date. I have had an intense training on molecular biology as a requirement to develop the therapy. I have also gained a variety of in vitro skills ranging from transcriptional assays to co-culture of mouse and human primary cells as well as a range of cell lines. I have also gained in vivo experience including tumour mouse models where I gained various competences to be able to handle mice for experimental studies. As part of the induction statistical course I have gained knowledge and practical skills on how to apply the appropriate statistical tests to support my biological findings.
Apart from my experimental skills, I have also gained other skills such as organisational skills, time management, writing skills, presenting skills and working as part of a team and independently. I also anticipate learning how to fully generate the therapy using viral vectors as well killing assays to enable the therapeutic efficacy of the therapy to be assessed.
Have you attended, presented a poster or spoken at any conferences? Throughout my PhD studies I have attended the following conferences: British Society of Immunology Congress 2019, Liverpool, UK; Cancer Science Unit Conference, Southampton 2018 & 2019; Humanised Mouse Symposium at Cambridge University, UK 2018 & 2019; The Institute for Cancer Vaccines and Immunotherapy Conference at the Royal Society in London, UK.
I presented a poster and received first prize for my poster presentation for two consecutive years at the Cancer Science Unit Conference, Southampton 2018 & 2019. In order to attend the BSI congress 2019 in Liverpool I received a BSI bursary to cover most of the expenses.
Have you completed any additional training? I have gained additional training on structural biology by using a Small-angle X-ray scattering beamline at the European Synchrotron Radiation Facility in Grenoble, France.
Sheila Olendo Barasa (Queen Mary)
Cancer Inflammation and Immunology
 About me: My first degree was a BSc in Anatomical Sciences with Industrial Experience, which I completed at the University of Manchester. I went on to obtain an MSc in Cancer and Molecular and Cellular Biology at Queen Mary University of London.
About me: My first degree was a BSc in Anatomical Sciences with Industrial Experience, which I completed at the University of Manchester. I went on to obtain an MSc in Cancer and Molecular and Cellular Biology at Queen Mary University of London.
What is your PhD project and how did you choose it? My PhD focuses on the role of long noncoding RNAs and the Tumour Microenvironment in High Grade Serous Ovarian Carcinoma. I considered a variety of factors when choosing my project. I compared the novelty of the research questions between each of my rotations. I also considered the resources that I would have access to, the research skills that I would learn and the ratio of computational to wet lab work involved. Furthermore, I tried to get the closest match between my learning style and my potential supervisor’s project management style.
How did you find the mathematical/computational component of the DTP? Coming from a background with minimal bioinformatics knowledge and skills, the taught bioinformatics module offered at Southampton University as part of the MRC DTP programme provided me with a solid foundation and reinforced my understanding of bioinformatics techniques, which I used in my rotations.
Do you have any publications? I contributed to Workforce Diversity: Let’s talk about race. I also have a pending publication.
Have you attended, presented a poster or spoken at any conferences? I attended and presented a poster at the 2019 EMBO/EMBL The Non-coding Genome Symposium, 16th - 19th October, in Heidelberg, Germany.
What have you enjoyed the most and the least about your experience so far? I was extremely pleased to learn that the MRC DTP programme offered PhD internships in conjunction with other organisations/industrial partners, both in the UK and abroad. My hope is that the opportunity for more internships develops to the extent that it becomes embedded in the MRC DTP programme, as an optional component, especially given that this will enable us to make informed decisions about our careers and increase our employability prospects post-MRC DTP.
Annabelle Minton (Southampton)
Cancer Sciences Pathway
What is your PhD project? My PhD project is examining the role of B-Cell receptor driven Autophagy and Phagocytosis in Chronic Lymphocytic Leukaemia (CLL). I applied for this project as my undergraduate project was examining the role of Autophagic cell death following endocannabinoid treatment, so I was already familiar with the types of pathways and assays utilised, and was interested in expanding upon this background knowledge. I already had an MRes before applying for a PhD, which was a single project rather than rotation based, and my PhD funding was for 3.5 years on an allocated project. So I went straight into this project.
What skills have you developed so far? Recovery and handling of primary CLL cells, immunoblotting, immunoflurescence, flow cytometry, phagocytosis and endocytosis assay development, development of a high-throughput autophagy assay. I have also developed my public speaking at both external and internal conferences.
I expect to more fully develop my ability to work independently toward assay optimisation and development in the final year of my PhD, since I will be starting a new chapter examining antigen presentation in CLL. This will involve collaboration with another institution, and setting up a previously untried assay for use with a reporter T-Cell clone.
Do you have any publications? BCR signaling contributes to autophagy regulation in chronic lymphocytic leukemia. Leukemia 34, 640–644 (2020). https://doi.org/10.1038/s41375-019-0557-y. This paper contains many results generated from the first year of my PhD.
Have you attended, presented a poster or spoken at any conferences? I attended the Autophagy UK 2018 conference and the UK CLL Forum 2019, where I gave a 15 minute oral presentation.
What have you enjoyed the most and the least about your experience so far? I enjoyed the support given during the first year of my PhD project by our lab group, and the CLL group in general. We have regular meetings and opportunities to present, and I think this is really something that has been well supported throughout the PhD- the opportunity to gain experience presenting at both internal meetings / conferences and external conferences.
Stephen Murtough (Queen Mary)
Cancer Inflammation and Immunology
 About me: I am a pharmacist by trade who was lured by the bright lights of the laboratory, the excitement of research, and by a fascination with biology. Following a long-standing desire to pursue research, I completed an MSc in Cancer and Molecular and Cellular Biology at Barts Cancer Institute during the 2016/17 academic year, which not only built my base knowledge of molecular biology, the disease of cancer, and modern immunotherapies, but it also heightened my desire to pursue research at a higher level through a PhD. Having seen first-hand the brilliance of the scientists and expertise at Barts, it was a no-brainer to apply for their 4-year MRC-funded DTP. Additionally, having now completed my first rotation, I am determined more than ever to dedicate myself to these studies; the atmosphere in the medical school is contagious!
About me: I am a pharmacist by trade who was lured by the bright lights of the laboratory, the excitement of research, and by a fascination with biology. Following a long-standing desire to pursue research, I completed an MSc in Cancer and Molecular and Cellular Biology at Barts Cancer Institute during the 2016/17 academic year, which not only built my base knowledge of molecular biology, the disease of cancer, and modern immunotherapies, but it also heightened my desire to pursue research at a higher level through a PhD. Having seen first-hand the brilliance of the scientists and expertise at Barts, it was a no-brainer to apply for their 4-year MRC-funded DTP. Additionally, having now completed my first rotation, I am determined more than ever to dedicate myself to these studies; the atmosphere in the medical school is contagious!
How did you find the mathematical/computational component of the DTP? The Quantitative Cell Biology module at Southampton was a fantastic introduction to bioinformatics, coding, and R Studio. Previously, I had never analysed large datasets or attempted any coding, and so this was a completely new skillset for me to develop. It is evident that this is a must-have skill for the modern biologist in the era of ‘Big Data’, where datasets of ever-increasing size and complexity are becoming commonplace in the laboratory. Therefore, exposure to this expertise at Southampton throughout the beginning of the DTP has been a privilege, gifting us a necessary vital edge.
Do you have any publications? I have no publications to date; however, I have had two abstracts published in the Journal of Investigative Dermatology. These are as follows: iRhom2-mediated immune dysregulation: Impact on the skin and oesophagus, May 2019, Volume 139, Issue 5, Supplement, Page S78 (2nd Author); and Type 2 Immunity Linked to iRhom2 and Tylosis with Oesophageal Cancer, September 2019, Volume 139, Issue 9, Supplement, Page S287 (1st Author).
Have you attended, presented a poster or spoken at any conferences? In 2019, I attended the following conferences: Keystone Symposia: Skin Health and Disease: Immune, Epithelial and Microbiome Crosstalk, Hannover, Germany, April 8-11. Here, I presented both a poster and a short talk. Additionally, I received a travel grant from the ESDR to attend this conference for the value of €1000; 49th ESDR meeting, Bordeaux, France, 18-21 September. Here, I presented a poster as part of a poster walk; and William Harvey Day at Queen Mary University of London where I presented a poster.
Have you completed any additional training? I have obtained my animal licence, and I have also visited the Gilliet lab in Lausanne where I learnt how to perform the tape-stripping skin wounding protocol.
What have you enjoyed the most and the least about your experience so far? The most enjoyable aspect so far has been to delve into new exciting areas of biology and research. The least enjoyable aspect has been the flipside; that the rotations are short and you have to move on, even when progress in an interesting area is being made.
Sara Ferri (Southampton)
Institute of Human Development and Health
 About me: I studied Biomedical Engineering at Politecnico di Milano in Italy, where I obtained my BSc degree (2015) and MSc degree (2017). I then moved to Southampton to start my PhD and now I’m in my final year and I work in a multidisciplinary team between the Faculties of Medicine and Engineering at the University of Southampton, with a collaboration with the University of Oxford.
About me: I studied Biomedical Engineering at Politecnico di Milano in Italy, where I obtained my BSc degree (2015) and MSc degree (2017). I then moved to Southampton to start my PhD and now I’m in my final year and I work in a multidisciplinary team between the Faculties of Medicine and Engineering at the University of Southampton, with a collaboration with the University of Oxford.
What is your PhD project and how did you choose it? My PhD is about the development of a new treatment to enhance the bone fracture repair process. The aim of the project is to produce drug loaded microbubbles and to deliver them to a fractured bone. The drug is then released upon exposure to ultrasound in a non-invasive way. In this way, only the target site is exposed to the drug and off-site side effects are avoided. I chose this project because I’ve always been interested in bones, bone fractures and biomechanics.
What skills have you developed so far? Since I started my PhD almost three years ago, I’ve gained confidence in working in a lab and in trying to validate experimental data with computational ones and vice versa. I’ve always been an organised person, and my PhD has given me the chance to further improve my organisational skills and time management. I’ve also improved my abilities to communicate my research, pitching it to different audiences.
Have you attended any conferences or contributed to any publications? I am currently working on two papers. I have attended several internal and external conferences, including: Faculty of Medicine Conference 2018 (Southampton General Hospital, winner of ”Best Poster Prize”); Modelling and Experiments in Drug Delivery Systems (MEDDS 2018, University of Glasgow, oral presentation); Cardiff Institute for Tissue Engineering and Repair Annual Scientific Meeting (CITER 2018, Cardiff, oral presentation); VII Faculty of Engineering Conference (Southampton 2019, winner of ”Best Oral Presentation Prize”); 24th European Symposium on Ultrasound Contrast Imaging (Rotterdam 2019, poster); 19th International Symposium of ISTU (Barcelona 2019, poster); Acoustical Society of America Annual Meeting (San Diego 2019, oral presentation); 25th European Symposium on Ultrasound Contrast Imaging (Rotterdam 2020, poster).
Michaela Balderstone (Queen Mary)
Cancer Inflammation and Immunology
What is your PhD project? I am based in Dr James Whiteford’s group in the Centre for Microvascular Research within the WHRI. My PhD project is on angiogenesis (the development of new blood vessels from pre-existing vasculature) in the context of eye diseases, such as Diabetic Retinopathy. I am currently focusing on the protein-type tyrosine phosphatase receptor CD148, to determine whether by targeting this we could halt the disease progression in eye diseases where angiogenesis is a feature.
I enjoy working in my lab, it is a supportive and sociable environment.
Do you have any publications? I was involved in, and so am named on, the review 'Syndecan-3 in Inflammation and Angiogenesis.'
Have you attended, presented a poster or spoken at any conferences? In the last year I have attended two conferences. At the September 2019 British Society for Matrix Biology meeting “Cell Adhesion Networks in Health and Disease” I presented a poster on my latest research.
Have you completed any additional training? In November 2018 I completed the Home Office course to gain my personal license.
What have you enjoyed the most and the least about your experience so far? My favourite experience so far on the DTP has been meeting other members of my cohort in Southampton in February 2019 and learning more about them and their research. My least favourite experience was when I first started and attended the Christmas meet and greet in December 2018, as I had no idea what the programme was about nor who the other people in attendance were, so it was very disorientating. Fortunately, since attending the Southampton Away Day in February 2019, other aspects of the DTP were introduced to me through other students on the programme.
Caitlin Davies (Queen Mary)
Cancer Inflammation and Immunology
 What is your PhD project? My PhD project is entitled “Application of circulating tumour cell (CTC) analysis to monitor and predict prostate cancer response to Docetaxel.” So far, the project involves isolating CTCs from patient samples and performing number counts before, during and after treatment, as well as RNA/protein analysis to determine differential expression of genes involved in resistance - with the view to develop a non-invasive liquid biopsy method of monitoring and predicting treatment response in a patient specific manner. The company ANGLE plc are involved in my PhD project.
What is your PhD project? My PhD project is entitled “Application of circulating tumour cell (CTC) analysis to monitor and predict prostate cancer response to Docetaxel.” So far, the project involves isolating CTCs from patient samples and performing number counts before, during and after treatment, as well as RNA/protein analysis to determine differential expression of genes involved in resistance - with the view to develop a non-invasive liquid biopsy method of monitoring and predicting treatment response in a patient specific manner. The company ANGLE plc are involved in my PhD project.
How do you think having a relationship with a company already at this stage of your PhD will help your career development? It’s a great opportunity to gain understanding of scientific research outside of academia, and to forge new professional relationships with people outside of the BCI. I will hopefully learn new, translation techniques that I can’t learn elsewhere that will aid my own research.
What have you enjoyed most about your experience so far? I have most enjoyed learning and improving my skills as a scientist, and meeting new and interesting people.
Justina Then (Queen Mary)
Cancer Inflammation and Immunology
What is your PhD project? I am based in Dr James Whiteford’s group in the Centre for Microvascular Research at the WHRI and my PhD project currently is in arthritis investigating the mechanisms involved fibroblast invasion and destruction into cartilage and how to target these for therapeutic treatment focusing on the relationship between syndecan-4 and PTPRsigma.
What skills have you developed so far? I have developed a range of skills so far, ranging from laboratory based techniques such as adhesion and phagocytosis assays. I hope to further develop my skills in transfections and in vivo models.
What have you enjoyed most and least about your experience so far? So far, my favourite experience has been meeting the people on the programme and hearing about their research. My least favourite experience has probably been my transfer from one lab to another.
Cohort 2016
James Davies (Southampton)
Immunity and Infection Pathway
 What is your PhD project? My project is looking at “Understanding human epidermal Langerhans cell tolerogenic function using systems biology” and centres around skin-resident Langerhans cells (LC) and the critical role they perform to coordinate skin immune responses and maintain immune tolerance. Dysregulation of tolerance at the skin can contribute to disease, emphasising the importance to further understand the mechanisms underlying LC immune regulation.
What is your PhD project? My project is looking at “Understanding human epidermal Langerhans cell tolerogenic function using systems biology” and centres around skin-resident Langerhans cells (LC) and the critical role they perform to coordinate skin immune responses and maintain immune tolerance. Dysregulation of tolerance at the skin can contribute to disease, emphasising the importance to further understand the mechanisms underlying LC immune regulation.
What skills have you developed so far? I have developed skills in cell culture, processing human skin samples, flow cytometry and FACS, Drop-seq, molecular methods (cDNA library processing for sequencing and qPCR), bioinformatic analysis using R and mathematical modelling methods (signalling Petri nets). I aim to further expand my skills using mathematical modelling during the remainder of my PhD.
How did you find the mathematic/computational component of the DTP? The DTP introduced me to computational biology with the first-year quantitative cell biology module and bioinformatic rotation projects. This has subsequently become a major element of my PhD research project. I am therefore very grateful that the DTP has given me the opportunity to learn these new skills and explore and appreciate an area of biomedical science I may have not shown interest in previously.
Have you attended, presented a poster or spoken at any conferences?I have attended a variety of conferences during my time on the programme. Most recently I have attended the 16th International Workshop on Langerhans Cells (2019) where I gave an oral presentation (travel bursary awarded); the British Society for Investigative Dermatology Annual Meeting 2019 where I gave an oral presentation (travel bursary awarded); and the University of Southampton Medical and Health Research Conference 2019, where I won the award for the Best Oral Presentation.
Did you carry out any additional training? I was a Biotechnology YES Young Enterprise Scheme competitor in 2017, for which I was awarded a BBSRC sponsorship. I attended the In Silico Systems Biology Course in 2017 at the European Bioinformatics Institute (EMBL-EBI), Wellcome Genome Campus.
Do you have any publications? I am first author of a preprint entitled 'Single cell transcriptomic analysis indentifies Langerhans cells immunocompetency is critical for IDO1- dependent ability to induce tolerogenic T cells.' bioRxiv (2019).
Charlotte Smith (Queen Mary)
Cancer Inflammation and Immunology
 About me: Before commencing the MRC-DTP course I obtained my undergraduate degree in Biomedical Science from Oxford Brookes, a Masters in Cancer Therapeutics here at Barts and spent two years working as a researcher in the Institute of Cancer Research, London.
About me: Before commencing the MRC-DTP course I obtained my undergraduate degree in Biomedical Science from Oxford Brookes, a Masters in Cancer Therapeutics here at Barts and spent two years working as a researcher in the Institute of Cancer Research, London.
What is your PhD project and how did you choose it? I chose to continue my final rotation in Molecular Oncology at BCI in the Martin lab as my PhD project. In this project, we aimed to establish the precise impact of DNA repair loss and DNA damage on PD-L1 expression and ultimately sensitivity to immune checkpoint inhibitors, and how this can be exploited therapeutically. I chose this particular project not only because I am fascinated by the research, but also because I felt like I would work well within the supportive and knowledgeable team and within a diverse and friendly large department.
What skills have you learnt so far? I have developed many skills throughout my MRes rotational year, which I would not have otherwise gained. New skills include InCell imaging, cloning, FACS and Flow Cytometry, Confocal Microscopy, tumour digestion and establishing primary cell cultures. I also have built upon existing skills and have had the opportunity to share these with members of my teams.
As my PhD unfolds I will gain more experience, namely in Mass Spectrometry. I also have had the privilege of gaining a sponsored CASE studentship from Astra Zeneca, who are interested in developing my project, providing me with the opportunity to work with them alongside the BCI.
Have you attended, presented a poster or spoken at any conferences? I recently attended the Gordon Research Conference on DNA damage, mutation and cancer in Ventura, California USA. I presented a poster titled: 'Identification of DNA mismatch repair mutational signatures that predict response to immune checkpoint blockade.' I’m also attending an EACR conference on DNA damage responses and cancer in Cambridge in April 2020, where I’m also presenting my poster.
Jodie Ackland (Southampton)
Immunity and Infection Pathway
 About me: I initially studied Biology at Aberystwyth University and really enjoyed the immunology and microbiology modules, realising that immunity and infection was the field that really captured my interest. Time spent in the lab for my dissertation project really showed me that I loved doing research, so I decided to pursue a PhD, which led me to the MRes PhD at Southampton University.
About me: I initially studied Biology at Aberystwyth University and really enjoyed the immunology and microbiology modules, realising that immunity and infection was the field that really captured my interest. Time spent in the lab for my dissertation project really showed me that I loved doing research, so I decided to pursue a PhD, which led me to the MRes PhD at Southampton University.
What is your PhD project and how did you choose it? My PhD project is ‘Investigating Host Pathogen Interactions in the Asthmatic Airway.' This project was extremely appealing to me as it encompasses both fields of Infection and Immunity, which are where my interests lie. Southampton is renowned for both respiratory and translational research, which is of course important as at the end of the day, our research aims to improve patient outcomes and quality of life.
What skills have you developed so far? During this PhD I have developed a variety of laboratory techniques including primary human and bacterial cell culture, in vitro infection assays, flow cytometry, qPCR, FISH, ELISA, gentamicin protection assays and cell cytotoxicity/proliferation assays. I have also been able to build on the skills learnt during the computational taught module by analysing a RNASeq data set that I generated as part of my project.
Have you undertaken any additional training? I have had additional training on IRIDIS, the University of Southampton high performance computing system, which is required for analysis of large data sets.
How did you find the mathematic/computational component of the DTP? The computational component introduced me to big data sets and the different ways of analysing such data. This was incredibly useful as a huge component of my PhD was to generate and analyse an RNASeq data set.
Have you attended, presented a poster or spoken at any conferences? I have attended several internal and external conferences, most recently including: the ERS Lung Science Conference, Estoril, Portugal (March 2019) and the Southampton Medical & Health Research Conference, Southampton (June 2019) where I gave thematic poster presentations; and the ERS International Congress, Madrid, Spain (October 2019) where I gave a poster discussion presentation. I won the British Association for Lung Research (BALR) travel award twice to attend the ERS International Congress in 2017 and 2019.
Have you won any prizes for your presentations? I won the ‘Best Poster Presentation’ Prize at the Southampton MedConf 2019 and ‘Outstanding Poster Prize’ at the Network for Anti-Microbial Resistance and Infection Prevention festival 2019.
What have you enjoyed the most and the least about your experience so far? I have really enjoyed the opportunity to develop my skills as a scientist through a combination of the lab rotations, taught modules and additional training courses provided by the IPhD programme. The nature of the IPhD also allows you to be able to interact with your cohort and other research groups, which helps you to establish a professional network that is crucial going forward in your scientific career. In addition, the IPhD Away Days give an excellent opportunity for all students to present in a safe and friendly environment, which allows you to see the breadth of research your fellow students are undertaking.
Lauren Cutmore (Queen Mary)
Cancer Inflammation and Immunology
 What is your PhD project and how did you choose it? I continued with my third rotation for my PhD, entitled “Role of oncofetal glycosaminoglycans (OFGAG) for the immune privilege of pancreatic cancer,” which looked at developing a new CAR-T cell for the treatment of pancreatic cancer. During the rotation I isolated primary human T cells from blood, performed PCR and lots of molecular biology to clone the constructs into a plasmid and performed cytotoxicity assays.
What is your PhD project and how did you choose it? I continued with my third rotation for my PhD, entitled “Role of oncofetal glycosaminoglycans (OFGAG) for the immune privilege of pancreatic cancer,” which looked at developing a new CAR-T cell for the treatment of pancreatic cancer. During the rotation I isolated primary human T cells from blood, performed PCR and lots of molecular biology to clone the constructs into a plasmid and performed cytotoxicity assays.
It was really hard for me to choose as I enjoyed all of my projects, but I chose this one because I am really interested in CAR-T cell therapy and liked the translational theme of the project.
How did you find the mathematic/computational component of the DTP? I think the bioinformatics part of the course was really valuable to me as it has allowed me to have a better understanding of what is possible to find from large data sets that already exist. Using the skills I have developed, I have been able to do some work independently and I hope to develop my skills further to incorporate more bioinformatics into my project in the future.
Do you have any publications? Pancreatic Cancer UK Grand Challenge: Developments and challenges for effective CAR T cell therapy for pancreatic ductal adenocarcinoma. Pancreatology (2020) pii: S1424-3903(20)30042-9. PMID: 32173257; A Day in the Life: 6 Cancer Researchers in 6 Different Countries; Standing up for Science – A Voice of Young Science Workshop.
Have you attended, presented a poster or spoken at any conferences/meetings? I was selected to attend a Voice of the Future event as a member of the Biochemical Society where I was part of a televised question time style interview of members of a Government Select Committee (13th March 2018). I attended an Art and Science workshop held by the biochemistry society in collaboration with UAL (27th March 2018). I presented a poster at the London Pancreas Workshop in May 2018. I presented a poster at the BACR student conference in November 2018. I was invited to a Stand up for Science workshop in Manchester about science policy (April 2018). I am also a STEM ambassador and volunteer at the Centre of the Cell, working with school children learning about science.
Have you completed any additional training? I have got a Home office licence and I have completed phlebotomy training.
What have you enjoyed the most and the least about your experience so far? My experience of the DTP program has been really positive, I have met a lot of really great people and been allowed to develop a wide range of skills. I have really enjoyed the extra courses and teaching provided by the course, for example the media training day. One of the less enjoyable parts of the course was the first term when we had a lot of coursework and teaching in combination with the lab project, which made planning your time difficult. Overall I’m really happy I chose the DTP program at Barts and I am loving my PhD.
Cindy Jawahar (Southampton)
Centre for Cancer Immunology
 About me: I did my undergraduate degree at Queen Mary in Medical Genetics. I decided to apply for the Southampton MRC DTP Cancer Pathway, as I thoroughly enjoyed the immunology and cancer biology modules that were offered on my undergraduate course, and was intrigued by the field of cancer immunotherapy.
About me: I did my undergraduate degree at Queen Mary in Medical Genetics. I decided to apply for the Southampton MRC DTP Cancer Pathway, as I thoroughly enjoyed the immunology and cancer biology modules that were offered on my undergraduate course, and was intrigued by the field of cancer immunotherapy.
What is your PhD project and how did you choose it? My PhD project is entitled 'Vaccine Strategies to Target the Cancer Mutanome' and it explores how tumour-specific mutations can be exploited to develop more effective vaccine-based immunotherapeutic strategies. I chose to continue my first lab rotation for my PhD as I found the team supportive, the project fascinating and liked the balance between the dry and wet lab work involved.
What skills have you developed so far? I have learnt a variety of skills so far - molecular biology, PCR, tissue culture, epitope prediction, ELISpot, in vivo work, DNA and RNA transfections, nuclofection and FACS, basic bioinformatics and understanding of genomics and transcriptomics data, basic understanding of R, in vitro mRNA synthesis. In the future I hope to develop a better understanding of genomics and transcriptomics analysis and tumour dissociation. I have had some additional training on immunology and using the high performance computing system at Southampton.
How did you find the mathematic/computational component of the DTP? The computational component on the DTP has been very valuable. It has given me a greater understanding of large data sets. I have also had the opportunity to familiarise myself with various packages on R, which I am using in my PhD. This experience was extremely useful in introducing me to coding, and analysing genomics, transcriptomics and proteomics data. It was also extremely useful in helping me understand when to use various statistical tests.
Have you attended, presented a poster or spoken at any conferences? I have attended a variety of conferences throughout my time on the programme. I have delivered poster presentations at these conferences, including at the Southampton Medical and Health Research Conference (2017), the Cancer Sciences Unit Conference, the 2nd Annual Next-Gen Immuno-Oncology Congress and the Southampton Medical and Health Research Conference. I gave an oral presentation on Targeting mutanome in multiple myeloma at the 11th Cancer sciences Unit Conference. I won the prize for Best Poster at the Wessex Immunology Group Annual Spring Meeting in 2017.
Have you won any other awards? I was awarded Third Place at the Faculty of Medicine 3 Minute Thesis in 2019. I have also won an internship by the Academy of Medical Sciences to work with the UKRI Medical Research Council Policy Team.
Minal Patel (Queen Mary)
Cancer Inflammation and Immunology
 What is your PhD project and how did you choose it? I chose to continue my third rotation- “Identification and interpretation of pathogenic noncoding mutations in pancreatic cancer”- as my PhD project. I am currently using bioinformatics to look at publicly available ChiP-seq and matching RNA-seq data to find hotspots of pathogenic non-coding mutations in active gene regulatory regions. Not only is this a subject area I am very interested in, the PhD also involves learning new bioinformatics and laboratory techniques that I have not used before and will be useful in the future.
What is your PhD project and how did you choose it? I chose to continue my third rotation- “Identification and interpretation of pathogenic noncoding mutations in pancreatic cancer”- as my PhD project. I am currently using bioinformatics to look at publicly available ChiP-seq and matching RNA-seq data to find hotspots of pathogenic non-coding mutations in active gene regulatory regions. Not only is this a subject area I am very interested in, the PhD also involves learning new bioinformatics and laboratory techniques that I have not used before and will be useful in the future.
How did you find the mathematic/computational component of the DTP? The mathematic/computational component of the DTP programme and taking bioinformatics rotations has definitely helped me with the bioinformatics part of my PhD project.
Do you have any publications? I have contributed to two publications during my PhD: The Identification and Interpretation of cis-Regulatory Noncoding Mutations in Cancer (Review). High-Throughput (2018) 8(1). PMID: 30577431; Signatures of inflammation and impending multiple organ dysfunction in the hyperacute phase of trauma: A prospective cohort study. PLOS Medicine (2017) 15, 10. e1002694. PMID: 28715416
Have you attended, presented a poster or spoken at any conferences? I attended the National Cancer Research Institute conference in November 2018. I presented a poster at the Somatic Evolution and Tumour Microenvironment Symposium at the Francis Crick Institute on 2nd Dec 2019.
Did you carry out any additional training? I attended a course in Python programming language in October 2017.
What have you enjoyed the most and the least about your experience? Overall I’ve enjoyed the varied experience I have gained rotating through three different laboratories and meeting new people. I have least enjoyed presenting my work, however, this is necessary and good experience.
The DTP and PhD has been enjoyable and challenging. I have learnt/developed many transferable skills and new laboratory techniques with the support of my supervisors and fellow colleagues.
James Dickie (Southampton)
Centre for Cancer Immunology
 About me: After graduating from my BSc in Biomedical Science, I spent a couple of years working as a technician in the NHS to build up my confidence in the lab, before applying to Southampton. I knew I was interested in cancer immunology, but little more than that. Despite having no bioinformatic background, I also want to move into a career involving bioinformatics, so I have been learning by doing during my PhD. Now I have established myself as my group’s go-to Bioinformatician, constantly improving my skills through real-life problem solving.
About me: After graduating from my BSc in Biomedical Science, I spent a couple of years working as a technician in the NHS to build up my confidence in the lab, before applying to Southampton. I knew I was interested in cancer immunology, but little more than that. Despite having no bioinformatic background, I also want to move into a career involving bioinformatics, so I have been learning by doing during my PhD. Now I have established myself as my group’s go-to Bioinformatician, constantly improving my skills through real-life problem solving.
What is your PhD project and how did you choose it? My project is to identify novel cancer vaccine targets, which can be utilised in an oral cancer vaccine. I wasn’t sure what sort of project I’d work on when I started, but during the initial iPhD lab rotations I found a particular interest in cancer vaccines, and identified an approach I could take to employ bioinformatics to contribute to an ongoing project.
What skills have you learnt so far? Bioinformatics mostly, including all sorts of big data processing and analysis. I’ve also become a very confident presenter, happily speaking to groups and crowds about my work and my field. My science writing is markedly better than when I began, I can comprehend far more from fields both familiar and novel than before, and I’ve learned to work independently, forming my own ideas and acting upon them. A bit of everything really.
How did you find the mathematic/computational component of the DTP? The Quantitative Biology module at the University of Southampton was a good training in the basics of bioinformatics and data analysis. It sparked my interest and increased my confidence to steer my PhD studies into mathematical and computational biology, as an independent scientist.
What have you enjoyed most and least about your experience so far? Academic freedom has been amazing for me. Being free to create my own questions, and find my own way of answering them, is incredibly satisfying. On the flip side of that though, my least favourite part is the inevitable dead ends. When you spend weeks if not months working on an experiment only to find nothing of any value. It’s frustrating, but you just have to accept it as unavoidable.
Sarah Johnson (Queen Mary)
Cancer Inflammation and Immunology
 How did you choose your PhD project? I spent my third rotation in Sarah McClelland’s research group and have chosen to continue this project as my PhD, entitled 'Development of a system to induce specific monosomies.' Our lab works on the underlying biology of the highly disrupted genomes of cancer, focussing on how huge alterations in chromosome structure and number are acquired and tolerated. When progressing from the MRes to the PhD, and choosing which lab to return to, I was looking for a group with a strong publishing record, expertise in techniques I was keen to learn, with projects spanning fundamental cell biology to translational work. For me, the McClelland group not only offered this, but also offered a project which I find interesting and am excited to work on, within a great team.
How did you choose your PhD project? I spent my third rotation in Sarah McClelland’s research group and have chosen to continue this project as my PhD, entitled 'Development of a system to induce specific monosomies.' Our lab works on the underlying biology of the highly disrupted genomes of cancer, focussing on how huge alterations in chromosome structure and number are acquired and tolerated. When progressing from the MRes to the PhD, and choosing which lab to return to, I was looking for a group with a strong publishing record, expertise in techniques I was keen to learn, with projects spanning fundamental cell biology to translational work. For me, the McClelland group not only offered this, but also offered a project which I find interesting and am excited to work on, within a great team.
What skills did you develop during the rotations? Through the rotations, I developed varied lab skills from western blotting, cell culture, immunohistochemistry, molecular biology, microscopy, and experience with CRISPR/Cas9 systems. To complement this, the course is structured such that the taught modules run in parallel to rotations to give a core foundation in the theory of research methods and also computational biology. For me, the content on bioinformatics has been invaluable.
Do you have any publications? I have contributed to two review/comment publications during my PhD: Watching cancer cells evolve through chromosomal instability. Nature (2019) 570(7760):166-167. PMID: 31182831; The emerging links between chromosomal instability (CIN), metastasis, inflammation and tumour immunity. Molecular Cytogenetics (2019) 12: 17. PMID: 31114634
Have you attended, presented a poster or spoken at any conferences? At the beginning of my final year, I got the opportunity to travel to Paris for a conference where I gave a talk presenting my PhD work. This was made possible by a BCI travel bursary covering all expenses.
Do you have any additional comments about your overall experience? My experience of the course has been excellent throughout. The collaborative nature of the course between the two universities offers unique opportunities to benefit from expertise from leading researchers at both Queen Mary and Southampton. Then the course itself is structured so that the MRes gives you a strong foundation which sets you up in the best way to excel in your PhD research, both from the taught modules and your rotation projects which expose you to a wider set of laboratory techniques and put your research in context of the field. To anyone considering a PhD, I would 100% recommend this course.
Chiara Banas (Southampton)
Immunity and Infection Pathway
 What rotations did you undertake in your first year?
What rotations did you undertake in your first year?
My PhD comprises studies of human lung disease. For my first rotation project, I investigated the effect of double-stranded RNA on extracellular matrix deposition in an in vitro virus infection model of the airway mucosa. I chose this project as I was able to learn multiple wet lab techniques including human cell culture, transepithelial resistance, western blot, ELISA and immunofluorescence microscopy. I presented this project as a poster discussion (2-minute talk followed by poster presentation) at the end of Year 1 when I attended my first international conference, European Respiratory Society (ERS) International Congress 2017, in Paris, France. I received the ERS Young Scientist Award for outstanding submitted written abstract, which included free registration and €600 to cover travel and accommodation.
I chose to do a bioinformatics-based second rotation project, where I analysed the transcriptomic and epigenetic cytokine production by human skin Langerhans cells. I was able to utilise and further develop the bioinformatics skills I learned from the Quantitative Cell Biology module.
Following this, I knew I wanted my PhD to have elements of both wet lab techniques and bioinformatics in airway disease. I then decided to do my third rotation project on a subject that would then lead on to my PhD project. My third rotation project and PhD investigates the global response of mast cells by RNA sequencing. I was able to use my wet lab techniques to optimise the conditions for preparing the samples and studying mast cell responses. I am looking forward to using my bioinformatics skills to analyse the data from RNA sequencing.
My PhD project is entitled: Role of IL-33 on human mast cell responses to rhinovirus infection in asthma.
Have you attended, presented a poster or spoken at any conferences? I attended the European Respiratory Society (ERS) International Congress in Paris, France – 2017. I won an ERS Young Scientist Award for outstanding submitted written abstract.
I have presented a poster of my PhD at the ERS Lung Science Conference 2018 in Estoril, Portugal. I received an ERS travel bursary award for my submitted abstract to present in Portugal. The award included free registration, €600 and a mentorship lunch at the conference. I attended the European Mast Cell and Basophil Research Network (EMBRN) 2019 meeting in Uppsala, Sweden. I used my MRC conference allowance, along with €300 I won from the Biolegend’s junior investigator travel award.
How have you found your overall experience of the programme so far? Overall, the integrated PhD programme has given me plenty of support in my training and has helped to develop my skills. The opportunity to do an oral presentation in front of my peers at the Integrated PhD Away Day from year one has helped me to gain confidence in my communication skills. I found the Quantitative Cell Biology module extremely helpful as it allowed me to incorporate the bioinformatics skills into my PhD. Furthermore, I was able to attend career events and MRC seminars, which have been informative and enjoyable experiences.
Kalum Clayton (Southampton)
Immunity and Infection Pathway
 What did you choose as your PhD project? My PhD project is 'investigating the cross-talk between structural and immune components of human skin to understand pathogen sensing in cutaneous health and inflammation.' Deconvolution of the cutaneous transcriptome from healthy and inflammatory skin conditions to investigate host expression in response to altered immune and microbial contexts. We aim to use single-cell RNAseq of reconstituted epidermis models to identify bacterial responses in keratinocytes to understand mechanisms underlying atopic dermatitis and psoriasis arising from host-microbiome dysbiosis and cutaneous inflammation.
What did you choose as your PhD project? My PhD project is 'investigating the cross-talk between structural and immune components of human skin to understand pathogen sensing in cutaneous health and inflammation.' Deconvolution of the cutaneous transcriptome from healthy and inflammatory skin conditions to investigate host expression in response to altered immune and microbial contexts. We aim to use single-cell RNAseq of reconstituted epidermis models to identify bacterial responses in keratinocytes to understand mechanisms underlying atopic dermatitis and psoriasis arising from host-microbiome dysbiosis and cutaneous inflammation.
I chose this project because of the computational biology aspect allowing me to further develop my bioinformatics skills and generating my own data in the wet lab. I really enjoyed working with the Systems Immunology Group for two of my rotations where I developed a keen enthusiasm for skin biology and bioinformatics, and had some fun along the way!
What skills have you developed so far? My undergrad background was molecular microbiology so my PhD project has really given me the opportunity to add human tissue culture such as developing reconstituted epidermal models and host-microbe co-cultures, and bioinformatics skills, such as coding, standard bulk and single-cell sequencing pipelines, and machine learning to my CV.
Do you have any publications?
Clayton K, Vallejo AF, Sirvent S, Davies J, Porter G, Lim FL, Ardern-Jones M, Polak ME. (2019) Machine learning applied to atopic dermatitis transcriptome reveals distinct therapy dependent modification of the keratinocyte immunophenotype. MedRxiv
Clayton K, Vallejo AF, Davies J, Sirvent S, Polak ME. (2017) Langerhans Cells-Programmed by the Epidermis. Frontiers in Immunology 8:1676. Review
Clayton K, Polak ME, Woelk CH, Elkington P. (2017) Gene Expression Signatures in Tuberculosis Have Greater Overlap with Autoimmune Diseases Than with Infectious Diseases. American Journal of Respiratory and Critical Care Medicine 196(5):655-656
Have you attended, presented a poster or spoken at any conferences? In 2017 I attended the Wessex Immunology Group Annual Conference (short talk presented); Faculty of Medicine Conference (poster presented). In 2018 I attended the British Society for Investigative Dermatology (poster presented); Faculty of Medicine Conference (poster presented); Wessex Immunology Group Annual Conference (short talk presented); Unilever Microbiology Symposium (short talk presented). In 2019 I attended the British Society for Investigative Dermatology (talk presented); Faculty of Medicine Conference (talk presented).
How have you found your overall experience of the programme so far? I have really enjoyed my PhD, which I think is mainly down to choosing to do a DTP with an MRes at the start. This allowed me to get a feel for different research projects and groups, where I could develop a project with a PI and group I knew I would enjoy working with. Doing bioinformatics was really out of my comfort zone at the start but I have developed an exciting project with some good results and useful skills that should come in handy for a career in research!
MRC iCASE
Cohort 2021
 Marcos Burger Ramos (Queen Mary)
Marcos Burger Ramos (Queen Mary)
Cancer Inflammation and Immunology
What is your PhD project? Broadly speaking, my PhD project with Dr Oliver Pearce is about targeting the extracellular matrix (ECM) in order to increase immune infiltration in triple negative breast cancer (TNBC). Like many other solid tumours, this subtype of breast cancer often shows poor cancer immunity which renders otherwise promising immunotherapies, such as CAR-T cell therapy, ineffective. One of the main barriers to immune infiltration is a very dense tumour ECM. With my PhD project, we will use tumour colonizing bacteria designed by Neobe Therapeutics to degrade certain ECM molecules in a decellularized tissue model of TNBC, and characterize their effect on CAR-T cell trafficking and cytotoxicity in this model.
What skills have you developed so far, and what skills do you hope to develop? Given that I only started about a month ago the list of skills I have developed as part of this PhD is not very long. However, it was good to do some bacterial work again as I had forgotten most of the practical techniques used for bacterial culture! One technique I am particularly looking forward to is live imaging of the CAR-T cells and the bacteria in my TNBC tissues which sounds quite complex but I think having videos of how these cells move through the tissues and potentially even how the bacteria express their payload will be incredibly interesting.
How do you think having a relationship with a company already at this stage of your PhD will help your career development? I think the intersection of academia and industry is an incredibly exciting space to be working in and being able to learn about the regulatory guidelines research in industry has to adhere to gives scientists a much better understanding of how their work can be translated and incorporated into patient care. For me, I think learning about these things this early on will help me a lot in producing impactful work and will also give me a much clearer picture of what type of research I’d like to do after my PhD.
What have you enjoyed most and least about your experience so far? What I’ve enjoyed the most so far is definitely how welcoming and helpful everyone is in the lab! It’s been very nice settling in when there are loads of people around to help show where things are etc. I’ve also really enjoyed my practical work with Neobe Therapeutics and hearing about the co-founders’ experiences starting a biotech start up in London. Something I maybe haven’t enjoyed as much is the slightly slower pace of my PhD in the first month but actually this has been quite useful to do all my background reading and make sure I have a rough plan for the next couple of months so it really isn’t a bad thing after all!
Cohort 2020
Matthew Milton (Southampton)
Immunity and Infection Pathway
 What is your PhD project? I have just started working towards my PhD project entitled ‘Investigating the functional role of CD1 expression and defining protective and pathogenic CD1c-immunity in tuberculosis’. With Mycobacterium tuberculosis infecting nearly a quarter of the world’s population and claiming 1.4 million lives each year, there is a desperate need for a new vaccine. For my project I am investigating specific subsets of T cells which we hypothesise could be important for protection against Mycobacterium tuberculosis infection. We hope that the findings from the project will inform the development of future TB vaccines.
What is your PhD project? I have just started working towards my PhD project entitled ‘Investigating the functional role of CD1 expression and defining protective and pathogenic CD1c-immunity in tuberculosis’. With Mycobacterium tuberculosis infecting nearly a quarter of the world’s population and claiming 1.4 million lives each year, there is a desperate need for a new vaccine. For my project I am investigating specific subsets of T cells which we hypothesise could be important for protection against Mycobacterium tuberculosis infection. We hope that the findings from the project will inform the development of future TB vaccines.
What skills have you developed so far, and what skills do you hope to develop? Looking back, I have learnt lots over the course of the iPhD so far. I have had the opportunity to gain skills and experience in everything from tissue culture through to bioinformatic analysis. I hope to soon get the training required to work in containment level 3 laboratories and learn how to conduct co-culture experiments incorporating live Mycobacterium tuberculosis.
How have you found the mathematic/computational component of the DTP? Having never even attempted coding, I thought that I would find the bioinformatics and computing elements of the iPhD course the most difficult. However, from taught content of the iPhD course along with being fortunate enough to have completed a bioinformatics rotation project, my skills and knowledge of bioinformatics have grown enormously. Thanks to what I have learnt, I expect bioinformatic analysis to form a valuable component of my PhD project.
What have you enjoyed most and least about your experience so far? Despite the rotation projects I have completed being spread across multiple different fields of research, they all provided an opportunity for me to gain valuable skills and experience which I am now using for my PhD project. However, completing three different rotation projects is a little like starting three new jobs in the same year, making the first year of the iPhD course quite intensive.
 Dillon Popat (Queen Mary)
Dillon Popat (Queen Mary)
Cancer Inflammation and Immunology
What is your PhD project focused on and how did you choose it? My iCase PhD involves the development of new drugs for the treatment of ACTH secreting tumours focussing specifically on the unique G protein-coupled receptor (GPCR), MC2R, and its accessory protein, MRAP. The MC2R/MRAP complex is involved in the binding of ACTH and post activation leads to the production of the stress hormone, cortisol, from the adrenal gland. High levels of cortisol reduce the quality of life of the patient and worsen the prognosis of neuroendocrine tumours. We are working with a company called OMass therapeutics who specialise in native mass spectrometry for drug development of GPCRs.
I chose this project so I would be able to work with a group who works very translationally. We use advanced in vitro and in vivo models and have direct links to the clinic and industry, this allow us to bring our research from benchwork to bedside. Working with industry means I will be able to experience various research environments and understand small and large-scale industrial research projects.
My project will allow me to use a variety of tools and work across disciplines, from molecular biology techniques to physical chemistry and bioinformatics.
What skills have you developed so far? So far, I have developed various skills in molecular and cell biology, including several biosensor assays and techniques to study GPCRs which are the biggest pharmacological targets. I have also started to use advanced computational modelling techniques like Alphafold.
What have you enjoyed most and least about your experience so far? I have most enjoyed working in an incredibly supportive department at the William Harvey Research Institute's Centre for Endocrinology, the people around me have made me a better scientist and have always encouraged new ideas.
Covid made it hard to initially start work in the lab and start to meet other people in the department, but now things have started to open we have been able to be more social.
Cohort 2019
Sahar Farag (Southampton)
Immunity and Infection Pathway
 What is your PhD project? My iCASE PhD project is investigating the immune response to tuberculosis, which represents a serious worldwide human health threat. I am studying the functional effect of CD1c responsive T cells in human Tuberculosis (TB). CD1c belongs to a family of lipid antigen presenting molecules that can present self- and foreign lipids to T cells, and thus initiate an immune response. Understanding the populations of T cells and mechanisms involved in fighting TB are essential for the development of effective vaccines and therapies. My project puts to use tetramers generated in-house to identify and isolate specific populations of T cells via flow cytometry; as well as traditional 2D cell culture and a 3D system to better understand the host-pathogen interaction. My PhD being an iCASE means that I also have an Industry Supervisor and will have the exciting opportunity to complete a placement at Immunocore (Abingdon, Oxford) where I’ll gain insight into what it’s like working in industry as well as be able to learn techniques and gain the most from their expertise to further my project.
What is your PhD project? My iCASE PhD project is investigating the immune response to tuberculosis, which represents a serious worldwide human health threat. I am studying the functional effect of CD1c responsive T cells in human Tuberculosis (TB). CD1c belongs to a family of lipid antigen presenting molecules that can present self- and foreign lipids to T cells, and thus initiate an immune response. Understanding the populations of T cells and mechanisms involved in fighting TB are essential for the development of effective vaccines and therapies. My project puts to use tetramers generated in-house to identify and isolate specific populations of T cells via flow cytometry; as well as traditional 2D cell culture and a 3D system to better understand the host-pathogen interaction. My PhD being an iCASE means that I also have an Industry Supervisor and will have the exciting opportunity to complete a placement at Immunocore (Abingdon, Oxford) where I’ll gain insight into what it’s like working in industry as well as be able to learn techniques and gain the most from their expertise to further my project.
What skills have you developed so far, and what skills do you hope to develop? Prior to starting this PhD I had no experience in flow cytometry. I only had theoretical knowledge in its principles; however, its application is very different, and I can now design FACS experiments, use the FACSAria confidently and analyse subsequent data using FlowJo. I have also learnt how to generate functional tetramers from protein inclusion bodies grown in bacteria, which is a cornerstone of my project. Moving forward, I hope to build on my ability to carry out manual compensation when setting up flow cytometry experiments; as well as learn more effective methods of cloning T cells.
How have you found the mathematic/computational component of the DTP? I have recently completed an introductory course on RNA-seq which uses Kallisto and R. Whilst my project does not necessarily include bioinformatics at the moment, it was really interesting to learn about how bioinformatics analyses are carried out, and be able to better understand published articles that include data derived from bioinformatics. It would be useful to complete a more in-depth course and possibly incorporate it in my project.
What have you enjoyed most and least about your experience so far? I have enjoyed learning completely new skills and techniques that I previously hadn’t been exposed to. I am also glad to be part of the group that I am in. As is the nature of science, not all experimental methods work which can be a little frustrating, but there are always other options to explore.
Sarah Hindle (Queen Mary)
Cancer Inflammation and Immunology
What is your PhD project and how did you choose it? My PhD project is based at the Blizard Institute and is supervised by Dr John Connelly. My research focuses on developing a vascularised and immune responsive in-vitro skin model using 3D bio-printing. I chose this project because I was interested in broadening my knowledge of 3D culture models and I was excited by the potential impacts of this research on the fundamental understanding of skin biology, in the discovery and testing of therapeutics and in the engineering of tissues for regenerative medicine purposes. Additionally, my project is an industrial CASE studentship, which means that I have an industrial partner, Kirkstall Ltd. Towards the end of my PhD I will be conducting a 3-month placement within their Research and Development team. During this placement I will gain an insight into the biotechnology industry and gain knowledge about fluidic cell culture systems and their applications for different tissue and disease models.
What skills have you developed so far? So far during my PhD I have learned to print in-vitro vascularised skin models using the 3D-bioprinter and have further developed my skills in adherent- and suspension-cell tissue culture, immunohistochemistry, confocal and epifluorescence imaging and data analysis.
Cohort 2018
Henry Gerdes (Queen Mary)
Cancer Inflammation and Immunology
 What is your PhD project and how did you choose it? My project aims to investigate the mechanisms which sensitise patients to Aurora B inhibitors in acute myeloid leukaemia using proteomic and systems biology approaches. I then identify predictive biomarkers of patient response to Aurora B inhibitors from the data generated using these techniques. I chose this project because it utilises and develops techniques which will improve precision medicine for cancer treatment. In addition it is also a great opportunity to gain experience in both academia and industry in a field I am passionate about.
What is your PhD project and how did you choose it? My project aims to investigate the mechanisms which sensitise patients to Aurora B inhibitors in acute myeloid leukaemia using proteomic and systems biology approaches. I then identify predictive biomarkers of patient response to Aurora B inhibitors from the data generated using these techniques. I chose this project because it utilises and develops techniques which will improve precision medicine for cancer treatment. In addition it is also a great opportunity to gain experience in both academia and industry in a field I am passionate about.
What skills have you developed so far? I have developed both laboratory and computational skills throughout my project. To generate proteomics data I use a technique called Liquid Chromatography Mass Spectrometry. This technique generates large datasets which I process and analyse using coding languages such as r and python. I have also been the president of the BCI PhD forum which has helped me further develop my leadership, organisation and communication skills.
How do you think having a relationship with a company already at this stage of your PhD will help your career development? Working closely with my company has given me more insight into how industrial research is organised and prioritised. This experience has allowed me to communicate with experts in my field and expanded my professional network. I will also be carrying out a work placement in Cambridge which will allow me to get first had experience working in industry.
What have you enjoyed most and least about your experience so far? I have enjoyed expanding my knowledge of bioinformatics throughout my project. Using these skills I have written small programs which automate a lot of my data analysis. Not only has this made me more productive it has produced tools which are useful for other members of my laboratory. The impact of COVID-19 on my project has been challenging. Luckily there have been a lot of resources available from Barts Cancer Institute and my industry sponsors which have helped me adapt my project goals in response to COVID-19.
Programme Information
Modules
Biofilms and Microbial Communities - University of Southampton
The module aims to provide an understanding of bacterial biofilms and the environmental, industrial and health care problems related to complex microbial consortia of societal importance. Students will learn to describe and explain the basis for biofilm development in nature and in chronic infections, as well as to understand and interpret the outputs of modern techniques in microbial biofilm research.
Biofilms and Microbial Communities Module Details, UoS - MRC DTP
Biological Therapies - Queen Mary University of London
This module will provide an understanding of the science behind the biological therapies of cancer, and an up-to-date review of the current status of preclinical and clinical trials. The module will also provide an insight into the ways that laboratory research is translated into clinical trials. By the end of the module you will be able to: Understand the basis of biological therapy of cancer; Understand the role of non-malignant cells and mediators in cancer growth and progression
Biomedical Parasitology - University of Southampton
The aim of this module is to introduce students to the main clinically relevant parasite classes, it will consider their lifecycles, the human/veterinary pathology caused and the treatment methods both of the primary and where applicable intermediate hosts and environment. It will give an understanding of vector borne disease and consider the interaction and evasion methods used by parasites in respect to the immune system and chemical control
Cancer Immunology - University of Southampton
The Cancer Immunology module provides an engaging and stimulating exploration of how immunology is modulated in disease, with a focus on cancer. It demonstrates the use of this knowledge to design novel immunotherapies. Alongside this immunology-specific knowledge, you will develop key skills in critical appraisal of previously published research, constructive discussion in small groups and oral presentation. The training covers the hot topics in Cancer Immunology. Each topic is taught by an academic working at the cutting edge in each research area. The topics span key immune and cellular processes:
- Vaccination and humoral immunity
- Antigen processing and presentation
- Dendritic cells
- T cell ontogeny/self-tolerance
- Autoimmunity and anti-tumour immunotherapy
Each week you will receive an initial seminar on the topic material, accompanied by a related journal paper that you will critique in a short written report for the following week. Then there follows an interactive discussion session where the paper is presented by one or more students. This interactive format provides an excellent learning experience, highly valued by students.
Cancer Immunology Module Details, UoS - MRC DTP
Communicable Disease Control - University of Southampton
Having successfully completed this module, you will be able to demonstrate knowledge and understanding of:
- Epidemiological methods to investigate and manage outbreaks.
- The principles of surveillance, the characteristics of different surveillance systems, their strengths and weaknesses and application to disease control.
- Application of communicable disease epidemiology to strategies for prevention and control leading to improvements in public health.
- The major methods for transmission of communicable disease.
Immunity and Infection - University of Southampton
The Immunity and Infection module has been designed to produce the next generation of leaders in immunity and infection research, some of the major research strengths of the University of Southampton. The module aims to provide you with an understanding of basic immunity and the underlying mechanisms of infection pathology. The course explores how disease mediated changes are managed and the different techniques and tools available to study them in detail. The course will enable you to develop your critical appraisal skills in the interpretation of published research. It will enhance your ability to present scientific data with focus and clarity.
The topics included in this module are:
- Innate Immunity and Mucosal Defence
- Key receptor interactions of the immune system: T-cell receptors, antibodies and integrins
- RNA Viruses, replication, manipulation, translational conundrums and avoiding innate immunity
- Bacterial infection in the airways: Tuberculosis, the host immune system and epidemiology
- Gene expression disease profiling, GWAS, RNA-seq, ChIP-seq, Proteomics - integration of Big Data in disease and inflammation.
Immunity & Infection Module Details, UoS - MRC DTP
Each week you will receive an initial seminar on the topic material, accompanied by a related journal paper that you will critique in a short written report for the following week. Then there follows an interactive discussion session where the paper is presented by one or more students. This interactive format provides an excellent learning experience, highly valued by students.
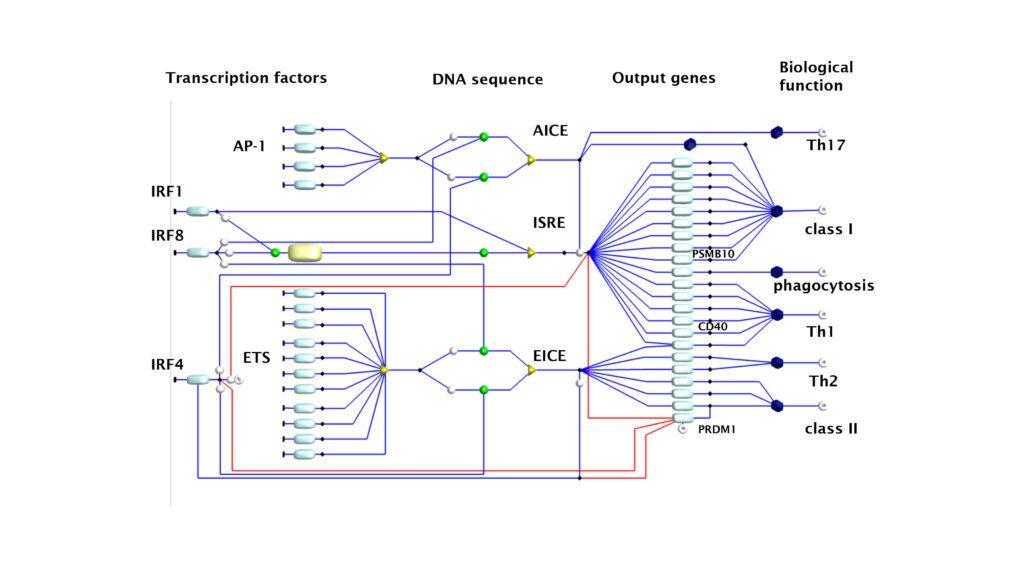
Methods and Analysis of Global Health Trends and Differentials - University of Southampton
The primary objective of this module is to provide an overview of the conceptual, methodological and empirical basis for quantifying levels of health in individuals and populations, including the construction of a range of different summary measures that combine information on mortality and non-fatal health outcomes. The course aims to give students an understanding of the technical basis for measurement in international work on population health; and to give students an appreciation of the uses and limitations of these methods in policy-making and priority-setting, particularly in developing countries.
Neurodegenerative Disease - University of Southampton
This module describes the neurobiology that underpins the aetiology and pathogenesis of neurodegenerative disease that has been a focus of intense and exciting research activity over the last several decades. The course is largely a research-led unit whereby the contributing staff will where possible, lecture on topics that are their own research interests.
Population & Reproductive Health - University of Southampton
The aim of this module is to enhance knowledge and understanding of the interrelationships between population characteristics, demographic, social and economic factors, and health. In this module we will take a global perspective to examine i) the influence of demographic patterns and change on health outcomes, and ii) the direct and indirect impacts on populations of changes in the burden of morbidity and mortality.
Population & Reproductive Health Module Details, UoS - MRC DTP
Quantitative Cell Biology - University of Southampton
Technological advances in nucleotide sequencing and proteomics are enabling scientists to study the molecular complexity of cellular systems, in biology and medicine, with unprecedented levels of detail. Whilst this greatly augments our ability to understand and treat disease, it also places new demands on biomedical scientists in how we generate, interpret and implement the very large, multivariate datasets that arise from such studies.
The Quantitative Cell Biology module is designed to allow you to begin to immerse yourself in this world, focusing on the practical use of the methods employed, so you can utilise their power to interrogate data in new and informative ways. Predominantly using "R" but with reference to other software tools, the module will provide you with an understanding of high-throughput “omics” technologies and the data they produce, allowing you to develop your skillset to analyse complex biomedical datasets. Specifically, over 5, one-day masterclasses, you will learn how to extract and manipulate these data, undertake unsupervised analysis, including clustering and dimensionality reduction, as well as supervised learning and classification and ending with deriving networks, pathways and models:
- R coding with formative assessment
- Extraction, normalizing and statistical analysis of data
- Transcriptomics – RNAseq analysis of multicellular tissues and single cells
- Deriving data visualisation networks from large datasets: clustering and dimensionality reduction
Quantitative Cell Biology Module Details, UoS - MRC DTP
Each masterclass will begin with a taught overview of the material, followed by hands-on sessions in the computer lab, with technical and mathematical support. Here you will learn to use various methods to analyse different data types and explore their power and potential in illuminating correlations and patterns in larger datasets. Collaborative working between students undertaking these sessions, and networking with more mature students in the PhD programme are positively encouraged as this builds confidence, skills transfer and rewarding working practices.
Examples of study output
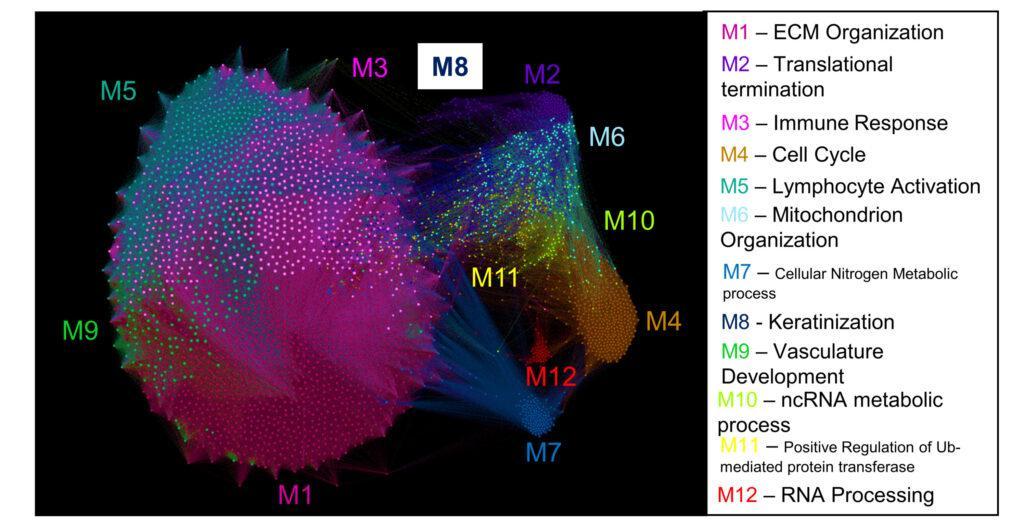
Transcriptional Networks in Cancer: The above Network map shows the relationship between different gene expression profiles measured by RNA-seq in multiple solid cancers. Genes with similar expression profiles represent functionally related “modules”, shown by the colour of each point (M1-12). This analysis identified four key modules associated with the tumour microenvironment (M1, M2, M3 & M5) and identified the enzyme NOX4 as a novel therapeutic target for inhibiting fibroblast activation in these tumours. Hanley et al., (2018). Targeting the Myofibroblastic Cancer-Associated Fibroblast Phenotype through Inhibition of NOX4. J Natl Cancer Inst. 110:109-120.
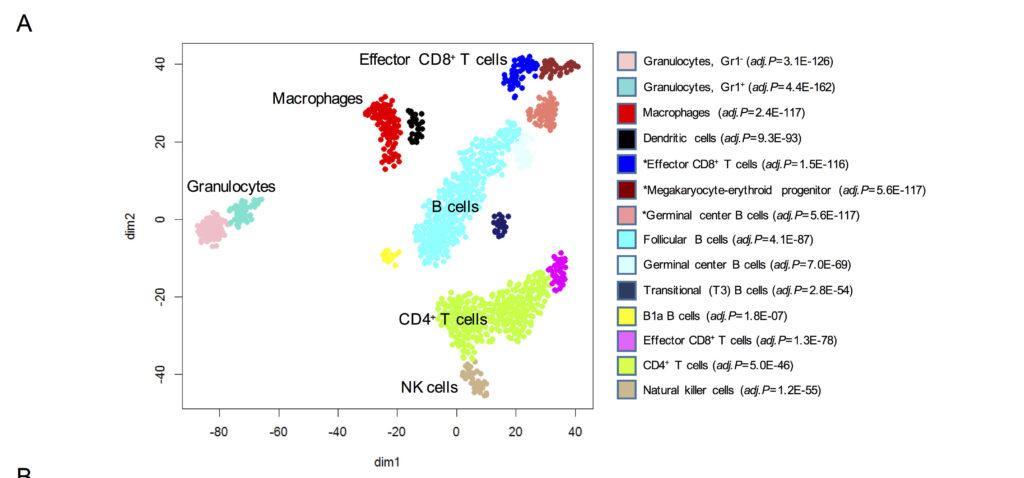
Single cell Gene Expression Profiling of lymphoma after treatment with Anti-CD20 and/or Anti-CD27 in mice. Single cell RNA-seq was performed on spleens, with t-SNE plots showing the individual immune effector subsets assigned by comparison to the co-expression atlas of the Immunological Genome Project. Each population is denoted by a different colour and those marked with an asterisk (∗) are proliferating.
Research Methods - Queen Mary University of London
The Research Methods module covers an introduction to statistical methods and tools, and their practical applications for student projects. The module includes a series of detailed lectures on the scientific basis of science research methods and allied skills.
The document below outlines the elements and goals of the module.
Systems Neuroscience - University of Southampton
This module seeks to expose students to research level studies in a number of areas related to the function of the nervous system, necessary to understand the pathophysiology of neurological conditions. The course will describe CNS development, and the structure and functions of neural cell populations; research into the mechanisms underlying learning & memory; and analyses of neural circuits controlling behaviour.
Annual Away Day at the University of Southampton
The MRC DTP Annual Away Day gives students from the University of Southampton and Queen Mary the opportunity to meet up and enjoy sharing their collective research experiences and challenges.
Students are empowered to help organise the day. The focus is on communication and interactions, where students present their research to their peers and supervisors, in a variety of formats. Students in their first year present one of their rotation projects, while students in their third or fourth year present data from their main PhD project. Moreover, students in the second year of the programme are tasked to present their entire PhD project to a non-specialist audience, in just three minutes with a single slide, which is very much enjoyed by everyone.
The Away Day is also an opportunity to gather some feedback on the programme. A lively forum is held that is attended by all participants. Here we discuss the programme openly together, with the aim of making positive changes to enhance the students’ research experiences and well-being and to talk about potential new opportunities with students.




Transferable Skills Training and Career Opportunities
Developing a broad range of research and transferable skills is one of the most important elements of studying for a higher research degree. In our programme, all the students are encouraged to develop their skills in a number of ways: practical research techniques in their chosen research topic, core transferable skills such as critical appraisal, data analysis and scientific writing. Presentation and critical analysis skills are developed through ongoing research work and taking part in research group meetings and journal clubs.
Students have access to a range of workshops and face-to-face training sessions. There are professional courses for graduate students, including e-learning resources, at the Doctoral College, University of Southampton. Generic skills training is encouraged, including team-working, time management, use of specialist software, effective communication and media training. Enterprise conferences and interactions with industry collaborators are organised by University staff. These include internships and placements, where students take a period of time, up to 3 months, to gain experience of complementary or parallel careers in science.
In today’s fluid and flexible jobs market, there is a greater need, than ever before, for students to have wide horizons and be cognisant of the career possibilities available to them. Regular workshops on opportunities and individual guidance are available to students through the Careers and Employability Service. The DTP Programme Team organises an annual conference showcasing Career Opportunities for PhD scientists. Professionals from different sectors come to talk about their career paths after a PhD in science. Contributors are suggested by the students themselves, in addition to the staff. Students have the opportunity to network and gather information on varied PhD level career opportunities.
Career Opportunities for PhD scientists is one of the most popular events in the programme calendar:
- How to Survive (and thrive) as an Academic in Industry
- How to start your own Biotech company - and have fun doing it
- A day in the life of a Commercial Translation Manager
- Personal Branding – Think outside the CV
- Behind the scenes at Nature Reviews - from bench to desk-side
- Science meets Law: From PhD to Patent Attorney
- Medical and Scientific Communications
- Interacting with Industry from a Research Perspective




Guidance for Applicants
Applications open in around November each year, with the programme commencing in September of the following year.
Writing an application for a DTP programme is much like applying for a job, but you’ll want to really highlight any research experience you’ve had, to show your commitment to this path. Use the tips listed in the drop-down boxes to help you tailor your CV and personal statement to good effect.
You will also find some advice on making a good impression at interview.
Please note, some of the links are available only to Queen Mary University of London or University of Southampton students; if you are studying currently at another university, be sure to make good use of the Careers Service where you are.
Eligibility
As this studentship is funded by the MRC, part of UKRI, please see UKRI's International eligibility criteria for funded studentships below.
Please note that this guidance is for students who will be recruited to start from the 2023/24 academic year.
To be classed as a Home student, candidates must meet the following criteria:
- Be a UK National (meeting residency requirements), or
- Have settled status, or
- Have pre-settled status (meeting residency requirements), or
- Have indefinite leave to remain or enter
UK National
The UK includes the United Kingdom and Islands (i.e. the Channel Islands and the Isle of Man).
In terms of residency requirements for UK nationals, for courses starting from 1 August 2021, candidates will continue to be eligible for home fee status as long as:
- they were living in the EEA or Switzerland on 31 December 2020, and have lived in the EEA, Switzerland, the UK or Gibraltar for at least the last 3 years before starting a course in the UK
- have lived continuously in the EEA, Switzerland, the UK or Gibraltar between 31 December 2020 and the start of the course
- the course starts before 1 January 2028
Eligibility on these grounds will only be available for courses starting up to seven years from the last day of the transition period (i.e. on 31 December 2027 at the latest).
Children of UK nationals will also be eligible for support on the same terms, even if they are not themselves UK nationals, as long as both the UK national and the child meet the conditions listed above.
This is following the announcement made by https://www.gov.uk/guidance/uk-nationals-in-the-eea-and-switzerland-access-to-higher-education-and-19-further-education
This will not apply to Irish nationals living in the UK and Ireland whose right to study and to access benefits and services will be preserved on a reciprocal basis for UK and Irish nationals under the Common Travel Area arrangement.
EU Settlement Scheme
EU, EEA or Swiss citizens can apply to the EU Settlement Scheme to continue living in the UK after 30 June 2021. If successful, applicants will get either settled or pre-settled status.
Settled status
Applicants will usually get settled status if they:
- started living in the UK by 31 December 2020
- lived in the UK for a continuous 5-year period (known as ‘continuous residence’)
Five years’ continuous residence means that for 5 years in a row they have been in the UK, the Channel Islands or the Isle of Man for at least 6 months in any 12-month period. The exceptions are:
- one period of up to 12 months for an important reason (for example, childbirth, serious illness, study, vocational training or an overseas work posting)
- compulsory military service of any length
- time spent abroad as a Crown servant, or as the family member of a Crown servant
- time spent abroad in the armed forces, or as the family member of someone in the armed forces
If they have settled status, they can spend up to 5 years in a row outside the UK without losing that status.
If they are a Swiss citizen, they and their family members can spend up to 4 years in a row outside the UK without losing their settled status.
Pre-settled status
If they do not have 5 years’ continuous residence when they apply, they will usually get pre-settled status. They must have started living in the UK by 31 December 2020.
Those with pre-settled status will qualify as a home student if they have 3 years residency in the UK/EEA/Gibraltar/Switzerland immediately before the start of their course.
It is then possible to apply to change this to settled status once they have got 5 years’ continuous residence. They must do this before the pre-settled status expires. They can stay in the UK for a further 5 years from the date they get pre-settled status.
If they have pre-settled status, they can spend up to 2 years in a row outside the UK without losing their status. They will need to maintain their continuous residence if they want to qualify for settled status.
Indefinite leave to remain (ILR) or Indefinite leave to enter (ILE)
Indefinite leave to enter or remain (ILR) are types of immigration status.
It is possible to continue to live in the UK without applying to the EU Settlement Scheme if they have indefinite leave to enter or remain in the UK. However, if they choose to apply (and meet all the other conditions), they will get ‘indefinite leave to remain under the EU Settlement Scheme’ - also known as settled status.
They can spend up to 2 years in a row outside the UK without losing their indefinite leave to enter or remain status.
International Student
If a candidate does not meet the criteria above, they would be classed as an International student.
Equality, Diversity and Inclusion
Queen Mary University of London and the University of Southampton are committed to making Equality, Diversity and Inclusion an integral component of University life. Both institutions hold Athena Swan Silver Awards and are Race Equality Charter signatories. We particularly welcome applications from those from minority ethnic groups, a disadvantaged background or having taken a career break. Flexible study is available and should be discussed at application (for UoS contact: Dr Sandrine Willaime-Morawek, s.willaime-morawek@soton.ac.uk; for Queen Mary contact Dr Katiuscia Bianchi, K.Bianchi@qmul.ac.uk). University benefits include onsite childcare facilities, state-of-the-art on-campus sports, arts and culture facilities and a full programme of events.
Writing your CV
-
Use headings well to highlight the most recent and relevant experience you’ve had
-
Provide well-chosen detail to give more information about the research projects you have undertaken at undergraduate, and possibly Masters, level
-
Split out your research skills - e.g. handling lots of information, lab skills, coding or data skills, project management skills: these are the skills that are really important in a PhD
-
Demonstrate a commitment to ongoing self-development, an important part of many PhD programmes, by showing what research methods or other training you have taken
-
Show how you have contributed to your current degree programme - e.g. organising department events such as journal clubs or something social - a PhD programme isn’t all research!
Useful links:
Applying for a PhD - The Careers Group
PhD Application Tips - Jobs.ac.uk
Writing your Personal Statement
- Use this structure to create at least three good paragraphs:
- Why are you interested in this PhD programme?
- Why do you want to come to Queen Mary or University of Southampton?
- Why do you think they should be interested in having you on their programme? This is where you get to mention some of the great things you have on your CV.
- Demonstrate the research you’ve done into doing a PhD, and into this programme. For example, mention that you’ve spoken to existing PhD students about their experiences, or how you’ve looked at some of the researchers’ work online.
- It’s important to be aware that moving into academia after a PhD is not guaranteed. Use your personal statement to show how becoming a really effective researcher with excellent transferable skills is important to you.
- Use enthusiastic and active language (‘developed’, ‘created’, ‘initiated’, ‘learned’) in your statement.
- If you can, tie your personal statement to what the PhD programme is specifically looking for. For example, if they are asking for ‘motivated students that can handle a lot of data with attention to detail,' explicitly provide examples of where you have demonstrated motivation, and where you have been successful in spotting errors or checking your work.
Useful links:
Queen Mary Careers CV support, including links to creating good cover letters
Tips for your interview
Congratulations on getting an interview!
- Deal with your nerves. Some people find that going for a walk, taking deep breaths, or having a reward ready for after an interview can be really good. The best thing though, is to have a practice!
- Have your application documents, and the programme specification, to hand during your interview so you can refer to bits of it if necessary.
- Have some really clear examples of the key skills required for the PhD programme ready: use Context, Action, Result as a structure for your examples. Or think in 3s - ‘There are three reasons I want to embark on this programme. Firstly….’. Practice!
- Don’t worry if you can’t immediately think of an answer. Perhaps ask the interviewer to repeat the question, or answer the part of it you can – acknowledging it’s not the whole answer. Or, tell them how you would find out: problem-solving is a big part of a PhD.
- If you are asked to do a presentation, then rehearse it to make sure of your timings and try to ensure you show off the most impressive parts of your research. Try to present a logical, clear story of what you did, how, and why. You can rehearse timings in PowerPoint and even record it to watch it back.
- Did we say practice already? Your careers service may well have spots available or online tools for getting interview feedback.
Useful links:
Queen Mary Careers Interview support, including how to book practice interviews and practicing online
Jobs.ac.uk support for interviews. Be aware that this advice is geared towards those looking for jobs after their PhDs; don’t be daunted, but take only what’s relevant to you!
Useful links
Jobs.ac.uk
Prospects
The Careers Group
University of Southampton